| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,506,815 – 1,506,909 |
| Length | 94 |
| Max. P | 0.500000 |

| Location | 1,506,815 – 1,506,909 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.31 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
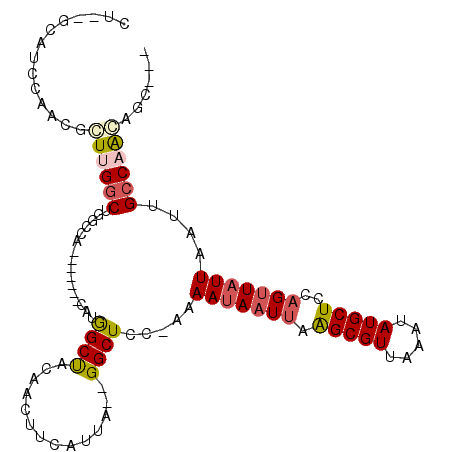
>3R_DroMel_CAF1 1506815 94 + 27905053 UU--GCAUCCAACGGUUGGCUGGCCA-------CCCGGCCACAACAUCAUUA--GGCUCC-AAAAUAAUUAGGCGUUAAAUAUGCUCCAGAUAUUAAUUGCCAGCC----- ..--.........((((((((((((.-------...)))))....(((....--(....)-..........(((((.....)))))...))).......)))))))----- ( -24.10) >DroVir_CAF1 10003 101 + 1 CU--GCAUCCAACGCUUGGCUGGUCA-------CAUGGCUACAACUUGAUUUGGGGCUCU-AAAAUAAUAAAGCGUUAAAUAUGCUCCAGUUAUUAAUUGGCCAGAGCUGC ..--.........(((((((..((..-------...(((..(((......)))..)))..-..((((((..(((((.....)))))...)))))).))..)))).)))... ( -23.70) >DroGri_CAF1 7362 101 + 1 CU--GCAACCAACGCUUGGCUGGCUA-------CAUGGCUACGUCUUCAUUUGGGGCUAC-AAAAUAAUUGGGCGUUAAAUAUGCUCCAGUUUUUAAUUGCCAACAGCGAU .(--((..((((.....((((((((.-------...))))).))).....))))(((...-.....((((((((((.....)))).)))))).......)))....))).. ( -28.76) >DroWil_CAF1 9319 105 + 1 UU--GCAUCCAACGCUUGGCUGCCUGGCUACGUCAUGGCUACGACUUCAUUA--GGCUCUUAAAAUAAUUAAGCGUUAAAUAUGCUCCAGUUAUUAAUUGCCAACAUCC-- ..--.........(.(((((.((((((....(((........)))....)))--)))......(((((((.(((((.....)))))..)))))))....))))))....-- ( -22.70) >DroAna_CAF1 6390 98 + 1 UU--GCAUCCAACGGUUGGCUGCCCA-------CCCAGCUACGACUUCAUUA--GGCUCC-AAAAUAAUUAAGCGUUAAAUAUGCUCCAGUUAUUAAUUGCCUACAGCA-C .(--((.......(((((((((....-------..))))..)))))....((--(((...-..(((((((.(((((.....)))))..)))))))....)))))..)))-. ( -23.90) >DroPer_CAF1 9251 102 + 1 CUGUGCAUAUAACGGUUGGCUGGCCACC-GCCACCUGGCCACAACUUCAUUA--GGCUCA-UAAAUAAUUAAGCGUUAAAUAUGCUCCAGUUAUUAAUUGCCAACA----- .............(((..(.((((....-)))).)..)))............--(((...-..(((((((.(((((.....)))))..)))))))....)))....----- ( -26.50) >consensus CU__GCAUCCAACGCUUGGCUGGCCA_______CAUGGCUACAACUUCAUUA__GGCUCC_AAAAUAAUUAAGCGUUAAAUAUGCUCCAGUUAUUAAUUGCCAACAGC___ ..............((((((................((((..............)))).....(((((((.(((((.....)))))..)))))))....))))))...... (-12.86 = -13.31 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:23 2006