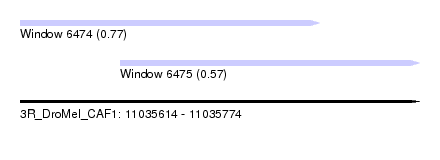
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,035,614 – 11,035,774 |
| Length | 160 |
| Max. P | 0.773079 |

| Location | 11,035,614 – 11,035,734 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -23.61 |
| Energy contribution | -23.03 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.773079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
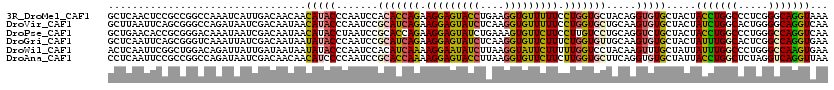
>3R_DroMel_CAF1 11035614 120 + 27905053 GCUCAACUCCGCCGGCCAAAUCAUUGACAACAACAUACCCAAUCCACACCAGAAGGAGUACCUGAAGGUGUUUUUCCUGGUGCUACAGGUGUGCUACUACCUGGCCCUCGGGCAGGUAAA (((....((((..(((((.....(((....)))((((((.......((((((.(((((((((....))))))))).)))))).....))))))........)))))..))))..)))... ( -35.10) >DroVir_CAF1 8536 120 + 1 GCUUAAUUCAGCGGGCCAGAUAAUCGACAAUAACAUACCCAAUCCGCAUCAGAAGGAGUAUCUCAAGGUGUUUUUCCUGGUGCUGCAAGUGUGCUACUAUCUGGCACUGGGGCAGGUCAA ((((..(((((...((((((((.......................(((((((.(((((((((....))))))))).))))))).(((....)))...)))))))).)))))..))))... ( -35.50) >DroPse_CAF1 9139 120 + 1 GCUGAACACCGCGGGACAAAUAAUCGACAAUAACAUACCUAAUCCGCACCAGAAGGAGUAUCUGAAAGUGUUCUUCCUUGUCCUGCAGGUCUGCUACUACCUGGCCCUGGGCCAGGUCAA ((.((...(.(((((((((......((((.....((((....(((.........))))))).......)))).....))))))))).).)).))....((((((((...))))))))... ( -35.70) >DroGri_CAF1 11036 120 + 1 GCUCAAUUCAGCGGGUCAAAUUAUCGACAAUAAUAUACCCAAUCCGCAUCAGAAGGAGUAUCUCAAGGUGUUCUUUCUGGUGUUGCAAGUGUGCUACUAUUUGGCACUCGGCCAGGUGAA ((.((.....((((((...(((((.....)))))..)))).....(((((((((((((((((....))))))))))))))))).))...)).))...((((((((.....)))))))).. ( -38.50) >DroWil_CAF1 20051 120 + 1 ACUCAAUUCGGCUGGACAGAUUAUUGAUAAUAAUAUACCCAAUCCACAUCAAAAGGAAUAUCUUAAGGUAUUCUUUUUGGUCCUACAAGUUUGCUAUUAUUUGGCCCUGGGCCAAGUGAA .........((((((....(((((.....)))))....)))......(((((((((((((((....)))))))))))))))...........))).((((((((((...)))))))))). ( -32.80) >DroAna_CAF1 8720 120 + 1 CCUCAAUUCCGCCGGCCAGAUAAUCGACAACAACAUCCCCAAUCCGCACCAAAAGGAGUACCUUAAGGUGUUCUUCUUGGUGCUUCAGGUGUGCUAUUACCUGGCUCUAGGUCAGGUUAA ..........((((((((((.........................(((((((.(((((((((....))))))))).))))))).(((((((......))))))).))).)))).)))... ( -38.90) >consensus GCUCAAUUCCGCGGGCCAAAUAAUCGACAAUAACAUACCCAAUCCGCACCAGAAGGAGUAUCUCAAGGUGUUCUUCCUGGUGCUGCAAGUGUGCUACUACCUGGCCCUGGGCCAGGUCAA .................................(((((.......(((((((.(((((((((....))))))))).))))))).....))))).....(((((((.....)))))))... (-23.61 = -23.03 + -0.58)

| Location | 11,035,654 – 11,035,774 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -24.10 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
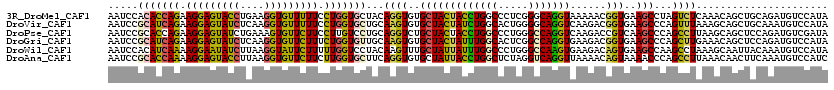
>3R_DroMel_CAF1 11035654 120 + 27905053 AAUCCACACCAGAAGGAGUACCUGAAGGUGUUUUUCCUGGUGCUACAGGUGUGCUACUACCUGGCCCUCGGGCAGGUAAAAACGGUGAAGCCUAGUCUCAAACAGCUGCAGAUGUCCAUA ......((((((.(((((((((....))))))))).))))))((.(((.(((((((.((((((.((....)))))))).....((.....)))))).....))).))).))......... ( -37.10) >DroVir_CAF1 8576 120 + 1 AAUCCGCAUCAGAAGGAGUAUCUCAAGGUGUUUUUCCUGGUGCUGCAAGUGUGCUACUAUCUGGCACUGGGGCAGGUCAAGACGGUGAAGCCCAGUUUAAAGCAGCUGCAAAUGUCCAUA .....(((((((.(((((((((....))))))))).)))))(((((....((((((.....))))))..((((...(((......))).))))........))))).))........... ( -35.60) >DroPse_CAF1 9179 120 + 1 AAUCCGCACCAGAAGGAGUAUCUGAAAGUGUUCUUCCUUGUCCUGCAGGUCUGCUACUACCUGGCCCUGGGCCAGGUCAAGACCGUCAAGCCCAGCCUUAAGCAGCUCCAGAUGUCGAUA .(((.(((...((((((.(((......)))))))))((.(..((((.(((((......((((((((...))))))))..)))))...(((......)))..))))..).)).))).))). ( -34.40) >DroGri_CAF1 11076 120 + 1 AAUCCGCAUCAGAAGGAGUAUCUCAAGGUGUUCUUUCUGGUGUUGCAAGUGUGCUACUAUUUGGCACUCGGCCAGGUGAAGACGGUGAAGCCCAGCUUGAAACAGCUCCAGAUGUCCAUA .....(((((((((((((((((....))))))))))))((.(((((((((((.((..((((((((.....)))))))).))))((.....))..)))))...)))).)).)))))..... ( -36.40) >DroWil_CAF1 20091 120 + 1 AAUCCACAUCAAAAGGAAUAUCUUAAGGUAUUCUUUUUGGUCCUACAAGUUUGCUAUUAUUUGGCCCUGGGCCAAGUGAAGACAGUGAAGCCAAGCCUAAAGCAAUUACAAAUGUCCAUA ....((((((((((((((((((....))))))))))))))).......((((....((((((((((...)))))))))))))).))).......((.....))................. ( -31.70) >DroAna_CAF1 8760 120 + 1 AAUCCGCACCAAAAGGAGUACCUUAAGGUGUUCUUCUUGGUGCUUCAGGUGUGCUAUUACCUGGCUCUAGGUCAGGUUAAAACAGUAAAACCCAGCCUUAAACAACUUCAAAUGUCCAUC .....(((((((.(((((((((....))))))))).))))))).(((((((......)))))))....((((..((((..........))))..))))...................... ( -35.50) >consensus AAUCCGCACCAGAAGGAGUAUCUCAAGGUGUUCUUCCUGGUGCUGCAAGUGUGCUACUACCUGGCCCUGGGCCAGGUCAAGACGGUGAAGCCCAGCCUAAAACAGCUCCAAAUGUCCAUA .....(((((((.(((((((((....))))))))).)))))))...((((..(((((((((((((.....)))))))......)))..)))...))))...................... (-24.10 = -22.80 + -1.30)
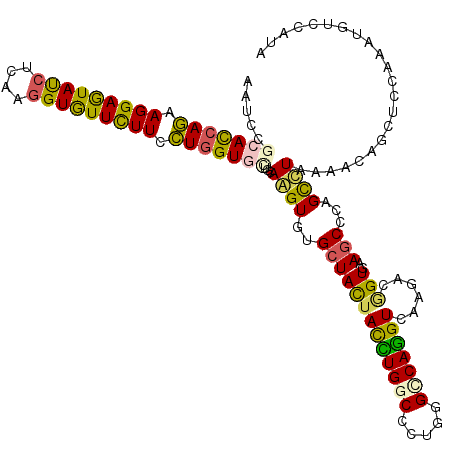
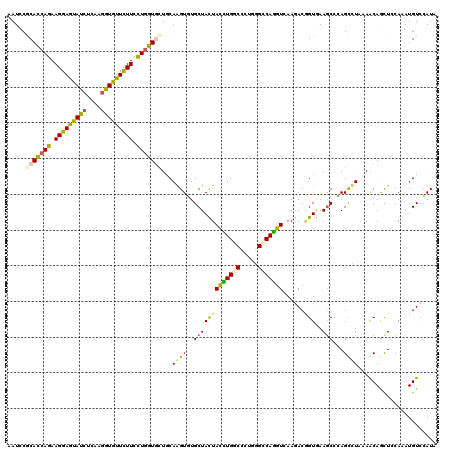
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:35 2006