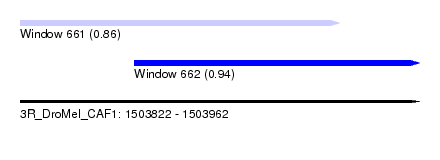
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,503,822 – 1,503,962 |
| Length | 140 |
| Max. P | 0.942598 |

| Location | 1,503,822 – 1,503,934 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.54 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -26.16 |
| Energy contribution | -26.33 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
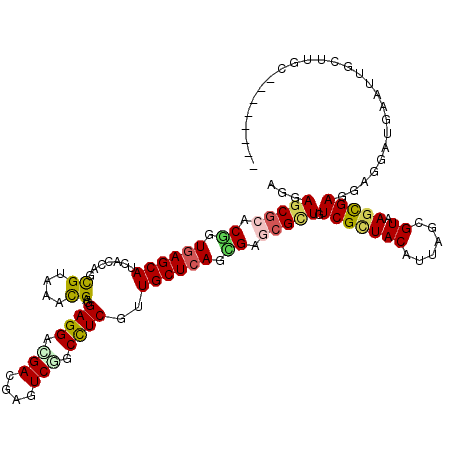
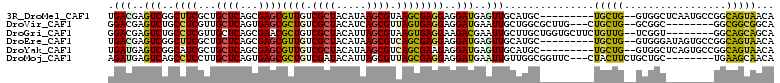
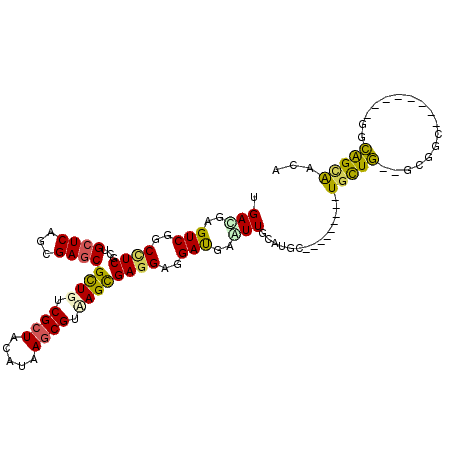
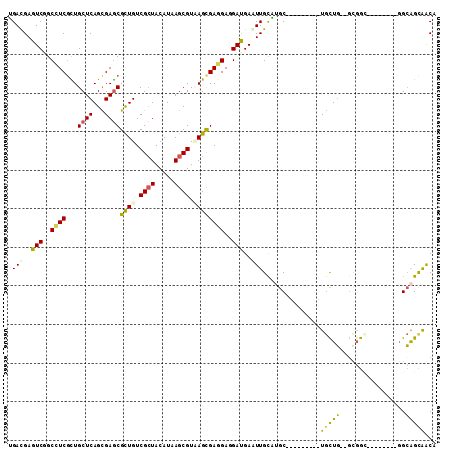
>3R_DroMel_CAF1 1503822 112 + 27905053 AGCAGCGCACGGUGAGCAUUACGAGCGUAAAUGAGGAGGAUGACGAGUCGGCUUCGCUGCUCAGCGAGCGUUGUCGCUACAUAAGCGUAAGCGAGGAGGAUGAGUUGCAUGC-------- .(((((((.((.((((((((((....))))..((((..(((.....)))..))))..)))))).)).)).((.(((((((......)).))))).))......)))))....-------- ( -39.60) >DroVir_CAF1 5609 117 + 1 AGGAGCGCACGGUGAGCAUCUCUAGCGUUAACGAGGAGGAGGACGAGUCUGCCUCGUUGCUCAGUGAGCGCUGUCGCUACAUCAGCGUUAGUGAGGAGGAUGAAUUGCUGGCGCUUG--- ..((((((.(((..(.((((.((.....((((((((....(((....))).))))))))((((.(((.(((((.........)))))))).)))).))))))..)..))))))))).--- ( -48.20) >DroGri_CAF1 4344 120 + 1 AGGAGCGCACGGUGAGCAUAAGCAGCGUCAACGAGGAGGAGGACGAGUCUGCCUCGUUGCUCAGCGAACGCUGUCGCUACAUUAGCGUAAGUGAGGAAGACGAAUUGCUUGCUGGUGCUU ..((((((.(((..((((.......(((((((((((....(((....))).))))))).((((.(..((((((.........))))))..)))))...))))...))))..))))))))) ( -49.70) >DroWil_CAF1 4139 112 + 1 AAGAGCGCACAGUGAGCAUAACCAGUGUCAAUGAGGAGGACGAUGAAUCGGCCUCGUUGCUAAGUGAGAGCUGUCGUUACAUAAGUGUCAGUGAGGAGAACGAACUGAUUGC-------- ...((((.(((((...(((.......(.((((((((....(((....))).)))))))))...)))...)))))))))........((((((..(.....)..))))))...-------- ( -30.50) >DroMoj_CAF1 4512 117 + 1 AAGAACGGACGGUGAGCAUCUCUAGCGUAAACGAGGAGGAAGAUGAGUCAGCCUCCUUGCUCAGUGAGCGCUGUCGAUACAUUAGCGUUAGCGAGGAGGAUGAAUUGUUGGCGGUUC--- ......((((.((.(((((((((..((....)).)))))........(((.((((((((((.....(((((((.........))))))))))))))))).)))..)))).)).))))--- ( -42.50) >DroPer_CAF1 4496 112 + 1 AGCAGCGCACUGUGAGCAUCACGAGCGUGAACGAGGAGGACGACGAGUCCGCUUCGUUGCUCAGCGAGCGCUGUCGCUACAUUAGCGUGAGCGAAGAAGAUGAACUGCUUGC-------- .(((((((.(..((((((((((....))))((((((.((((.....)))).))))))))))))..).))))))).((((...))))(..((((............))))..)-------- ( -46.60) >consensus AGGAGCGCACGGUGAGCAUCACCAGCGUAAACGAGGAGGACGACGAGUCGGCCUCGUUGCUCAGCGAGCGCUGUCGCUACAUUAGCGUAAGCGAGGAGGAUGAAUUGCUUGC________ ...(((((.((.((((((.......((....))..((((.(((....))).))))..)))))).)).))))).(((((((......)).))))).......................... (-26.16 = -26.33 + 0.17)

| Location | 1,503,862 – 1,503,962 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.32 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -20.53 |
| Energy contribution | -19.93 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1503862 100 + 27905053 UGACGAGUCGGCUUCGCUGCUCAGCGAGCGUUGUCGCUACAUAAGCGUAAGCGAGGAGGAUGAGUUGCAUGC---------UGCUG--GUGGCUCAAUGCCGGCAGUAACA .(((..(((..(((((((((((...)))).....((((.....))))..)))))))..)))..)))...(((---------(((((--(((......)))))))))))... ( -40.40) >DroVir_CAF1 5649 98 + 1 GGACGAGUCUGCCUCGUUGCUCAGUGAGCGCUGUCGCUACAUCAGCGUUAGUGAGGAGGAUGAAUUGCUGGCGCUUG---CUGCUG--GCGGC--------GGCGGCGGCA .((((((.....))))))((((...))))(((((((((.....((((((((..(..........)..)))))))).(---(((...--.))))--------))))))))). ( -44.80) >DroGri_CAF1 4384 101 + 1 GGACGAGUCUGCCUCGUUGCUCAGCGAACGCUGUCGCUACAUUAGCGUAAGUGAGGAAGACGAAUUGCUUGCUGGUGCUUCUGUUG--UCGGU--------GGCAGCAGCA ......((((.(((((((((.((((....))))..((((...)))))))).))))).))))........(((((.((((.(((...--.))).--------)))).))))) ( -39.20) >DroEre_CAF1 4724 100 + 1 UGACGAGUCGGCUUCGCUGCUCAGCGAGCGUUGUCGCUACAUAAGCGUCAGCGAGGAGGAUGAGUUGCAUGC---------UGCUG--GUGGGAUAGUGCCGGCAGUAACA .(((..(((..(((((((((((...))))...(.((((.....)))).))))))))..)))..)))...(((---------(((((--((........))))))))))... ( -42.00) >DroYak_CAF1 3728 100 + 1 UGAUGAGUCGGCAUCGCUGCUCAGCGAGCGUUGUCGCUACAUAAGCGUCAGCGAAGAGGAUGAGUUGCAUGC---------UGCUG--GUGGCUCAGUGCCGGCAGUAACA ......((((((((.((..(.((((...(((((.((((.....)))).)))))...((.(((.....))).)---------)))))--)..))...))))))))....... ( -42.10) >DroMoj_CAF1 4552 100 + 1 AGAUGAGUCAGCCUCCUUGCUCAGUGAGCGCUGUCGAUACAUUAGCGUUAGCGAGGAGGAUGAAUUGUUGGCGGUUC---CUACUUCUGCUGC--------UGAAGCAACA .(.((..((((((((((((((.....(((((((.........))))))))))))))))...........(((((...---......)))))))--------)))..)).). ( -35.50) >consensus UGACGAGUCGGCCUCGCUGCUCAGCGAGCGCUGUCGCUACAUAAGCGUAAGCGAGGAGGAUGAAUUGCAUGC_________UGCUG__GCGGC________GGCAGCAACA .(((..(((..((((...((((...))))((((.((((.....)))).))))))))..)))..)))...............(((((.................)))))... (-20.53 = -19.93 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:22 2006