
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,015,478 – 11,015,583 |
| Length | 105 |
| Max. P | 0.923505 |

| Location | 11,015,478 – 11,015,583 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -51.32 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
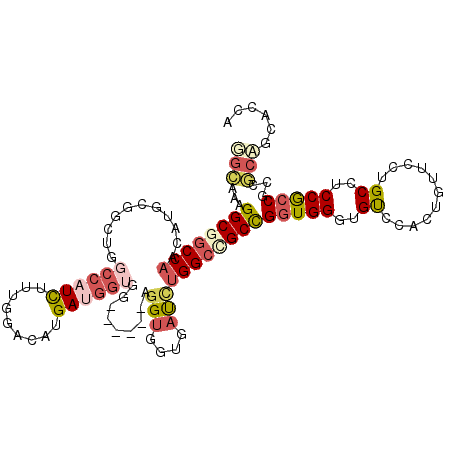
>3R_DroMel_CAF1 11015478 105 + 27905053 GGCAAAGGCGGCCAACAUGCGUCUGGCCCUUUACGGCCACGAUGG---------AGGUGGUGAUUUGGCUGCCGGUGGGUGUCCACUGUUUCUGCCUCCGCCGCCGCAGCAGCA .((...((((((........((((((((......))))).)))((---------(((..(.(((.(((.((((....)))).)))..))).)..)))))))))))...)).... ( -48.50) >DroVir_CAF1 127856 108 + 1 GGCAAAGGCGGCCAAUAUGCGCCUCGGCAUUUUUGGCCACGAUGGUGGUG------GCGAUGAUCUGGGAGCUGCUGGCUGUCCAUUAUUUCUGCCACCACCACCGCCGCCUCA (((...(((((((((.((((......))))..))))))..(.((((((((------(((..(((.(((((((.....))).))))..)))..))))))))))).)))))))... ( -53.20) >DroPse_CAF1 93692 114 + 1 UGUAAAGGCGGCCAACAUGCGGUUGGCCAUCUUUGGACAUGAUGGUGGAGGAGGAGGUGGGGAUCUGGCCGCUGGUGGUUGUCCUUUGUUCCUGCCUCCGCCGCCACAGCCCCA ...((((..(((((((.....)))))))..))))((...((.((((((.((((((((.((((((..(((((....)))))))))))....))).))))).))))))))...)). ( -54.50) >DroEre_CAF1 78529 105 + 1 GACAAAGGCGGCCAACAUGCGGUUGGCCCUCUAUGGCCAUGAUGG---------CGGUGGUGACUUGGCCGCCGGUGGGUGUCCACUGUUUCUGCCUCCGCCGCCGCAGCAGCA ..((.(((.(((((((.....))))))).))).))((..((...(---------((((((((....(((.(.((((((....))))))...).)))..)))))))))..)))). ( -47.70) >DroAna_CAF1 106877 108 + 1 GGCCAAGGCGGCCCAUAUGCGUCUGGCGAUUUUCGGACAUGAUGGUGG------CGGUGGAGACUUGGCAGCCGGUGGGUGCCCACUCUUCUUGCCACCACCGCCUCCACAACA ((((.....)))).......((((((......)))))).((.(((.((------((((((.....((((((..(((((....)))))....)))))))))))))).)))))... ( -49.50) >DroPer_CAF1 92002 114 + 1 UGUAAAGGCGGCCAACAUGCGGUUGGCCAUCUUUGGACAUGAUGGUGGAGGAGGAGGUGGGGAUCUGGCCGCUGGUGGUUGUCCUUUGUUCCUGCCUCCGCCGCCACAGCCCCA ...((((..(((((((.....)))))))..))))((...((.((((((.((((((((.((((((..(((((....)))))))))))....))).))))).))))))))...)). ( -54.50) >consensus GGCAAAGGCGGCCAACAUGCGGCUGGCCAUCUUUGGACAUGAUGGUGG______AGGUGGUGAUCUGGCCGCCGGUGGGUGUCCACUGUUCCUGCCUCCGCCGCCGCAGCACCA (((...((((((((...........((((((.........)))))).........(((....)))))))))))(((((..((...........))..)))))...)))...... (-21.62 = -22.57 + 0.95)

| Location | 11,015,478 – 11,015,583 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -20.53 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11015478 105 - 27905053 UGCUGCUGCGGCGGCGGAGGCAGAAACAGUGGACACCCACCGGCAGCCAAAUCACCACCU---------CCAUCGUGGCCGUAAAGGGCCAGACGCAUGUUGGCCGCCUUUGCC .(((((....)))))...((((((....((((....)))).(((.(((((..........---------.((((((((((......))))..))).)))))))).))))))))) ( -40.70) >DroVir_CAF1 127856 108 - 1 UGAGGCGGCGGUGGUGGUGGCAGAAAUAAUGGACAGCCAGCAGCUCCCAGAUCAUCGC------CACCACCAUCGUGGCCAAAAAUGCCGAGGCGCAUAUUGGCCGCCUUUGCC .(((((.((((((((((((((.((.....(((..(((.....))).))).....))))------))))))))))))((((((..((((......)))).))))))))))).... ( -54.10) >DroPse_CAF1 93692 114 - 1 UGGGGCUGUGGCGGCGGAGGCAGGAACAAAGGACAACCACCAGCGGCCAGAUCCCCACCUCCUCCUCCACCAUCAUGUCCAAAGAUGGCCAACCGCAUGUUGGCCGCCUUUACA ..(((((((((.((.(((((.(((......((....))......((........)).)))))))).)).))).)).))))((((.((((((((.....)))))))).))))... ( -45.20) >DroEre_CAF1 78529 105 - 1 UGCUGCUGCGGCGGCGGAGGCAGAAACAGUGGACACCCACCGGCGGCCAAGUCACCACCG---------CCAUCAUGGCCAUAGAGGGCCAACCGCAUGUUGGCCGCCUUUGUC .(((((....)))))((.(((...((((((((.........(((((...........)))---------))....(((((......))))).)))).)))).))).))...... ( -43.90) >DroAna_CAF1 106877 108 - 1 UGUUGUGGAGGCGGUGGUGGCAAGAAGAGUGGGCACCCACCGGCUGCCAAGUCUCCACCG------CCACCAUCAUGUCCGAAAAUCGCCAGACGCAUAUGGGCCGCCUUGGCC ....((.((((((((((((((.....(.((((....)))))((((....))))......)------))))).((((((.((....((....)))).)))))))))))))).)). ( -45.00) >DroPer_CAF1 92002 114 - 1 UGGGGCUGUGGCGGCGGAGGCAGGAACAAAGGACAACCACCAGCGGCCAGAUCCCCACCUCCUCCUCCACCAUCAUGUCCAAAGAUGGCCAACCGCAUGUUGGCCGCCUUUACA ..(((((((((.((.(((((.(((......((....))......((........)).)))))))).)).))).)).))))((((.((((((((.....)))))))).))))... ( -45.20) >consensus UGCGGCUGCGGCGGCGGAGGCAGAAACAAUGGACACCCACCAGCGGCCAAAUCACCACCU______CCACCAUCAUGGCCAAAAAUGGCCAACCGCAUGUUGGCCGCCUUUGCC ......(((((.(((((.(((........(((....)))......))).....................(((.((((..................)))).)))))))).))))) (-20.53 = -21.06 + 0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:18 2006