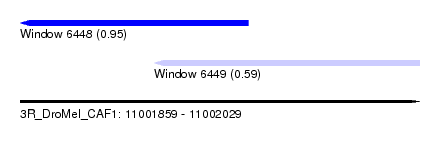
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,001,859 – 11,002,029 |
| Length | 170 |
| Max. P | 0.945673 |

| Location | 11,001,859 – 11,001,956 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.00 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11001859 97 - 27905053 UGCCCCUGGAGUACGAGCCAGAGGACAAGCUGCUGGCUGAGCUCAACAAGGGUAAGGAUAUCAAAGACAUUUUCUAAAUCCAA-------UGCCAGAC---A---CAAAA (((((.((((((.(.((((((((......)).))))))).))))..)).))))).((((.....(((.....)))..))))..-------........---.---..... ( -25.20) >DroPse_CAF1 78851 106 - 1 UGCCUUUGGAGUAUGAGCCAGAGGACAAGCUGCUGGCGGAGCUCAACAAGGGUAAGGACAAAAAAGACAUUUACUAAAAUCAAAAAACAAAUCCAAAC-CAA---AAAAA ((((((((((((....(((((((......)).)))))...))))..)))))))).(((..........((((....))))...........)))....-...---..... ( -22.50) >DroGri_CAF1 77421 91 - 1 UGCCUUUGGAAUACGAGCCAGAGGAUAAGCUGUUGACAGAACUCAACAAGGGUAAGGAUAUUAAAGACAUUUUCUAAAUACG----------UCCAAAAAA--------- ..(((((((........)))))))......((((((......)))))).......((((.....(((.....)))......)----------)))......--------- ( -18.20) >DroWil_CAF1 72406 104 - 1 UGCCUUUGGAAUACGAGCCAGAGGACAAGUUGCUGGCACAACUCAACAAGGGUAAGGAUAUCAAAGACAUUUUCUAAACUCAAAAA--A-AACCAAAA---AAAAAAAAA (((((((((........))))))).))(((((......)))))......((((..(((..............)))..)))).....--.-........---......... ( -16.94) >DroMoj_CAF1 86362 97 - 1 UGCCUUUGGAAUACGAGCCAGAGGAUAAGCUGUUGGCUGAACUCAACAAGGGUAAGGAUAUCAAAGACAUUUUCUAAAUACG----------UCAAAAAAAA---AAAAA ((((((((.......((((((.((.....)).))))))........))))))))...........(((.............)----------))........---..... ( -18.58) >DroPer_CAF1 76648 105 - 1 UGCCUUUGGAGUAUGAGCCAGAGGACAAGCUGCUGGCGGAGCUCAACAAGGGUAAGGACAAAAAAGACAUUUACUAAAAUCAAA-AACAAAUCCAAAC-CAA---AAAAA ((((((((((((....(((((((......)).)))))...))))..)))))))).(((..........((((....))))....-......)))....-...---..... ( -23.05) >consensus UGCCUUUGGAAUACGAGCCAGAGGACAAGCUGCUGGCAGAACUCAACAAGGGUAAGGAUAUCAAAGACAUUUUCUAAAUUCAA_______A_CCAAAA__AA___AAAAA ((((((((........(((((.((.....)).))))).........))))))))........................................................ (-16.16 = -16.00 + -0.17)

| Location | 11,001,916 – 11,002,029 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.73 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -23.58 |
| Energy contribution | -24.33 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11001916 113 - 27905053 UAUUAUC-----CACUUAGGUACCCAUCUCGCUAUGGUCUCUACUUGAGGGACAAGCCACUCACGCCCAACUCGCUGGUGCCCCUGGAGUACGAGCCAGAGGACAAGCUGCUGGCUGA .....((-----(.((((((((.((((......))))....)))))))))))..(((((..((.((....((((((.(((((....).)))).)))..))).....)))).))))).. ( -34.30) >DroVir_CAF1 97953 118 - 1 UAUUGUUUGUGCCUUUCAGAUAUCCAUCUCGCUAUGGUCUCUACUUGAGAGACAAGCCACUCACACCAAACUCUUUGGUGCCUUUGGAAUACGAGCCAGAGGAUAAGCUGUUGACUGA ................(((.(((((.(((.(((.((((((((.....))))))...(((....((((((.....))))))....)))....))))).))))))))..)))........ ( -30.80) >DroGri_CAF1 77472 118 - 1 UAUUAUUUAUGCGUUGCAGAUAUCCAUCUCGCUAUGGUCUCUACUUGAGAGACAAGCCACUCACACCAAACUCUUUGGUGCCUUUGGAAUACGAGCCAGAGGAUAAGCUGUUGACAGA ............(((((((.(((((.(((.(((.((((((((.....))))))...(((....((((((.....))))))....)))....))))).))))))))..))).))))... ( -34.00) >DroWil_CAF1 72470 105 - 1 -------------UUAUAGAUACCCAUCUCGCUAUGGUCUCUACUUGAGAGACAAGCCACUCACACCAAACUCCUUGGUGCCUUUGGAAUACGAGCCAGAGGACAAGUUGCUGGCACA -------------....((((....)))).(((...((((((.....)))))).(((.(((..((((((.....))))))(((((((........)))))))...))).))))))... ( -31.90) >DroMoj_CAF1 86419 107 - 1 -----------AUUUUCAGAUACCCAUCUCGCUAUGGUCUCUAUUUGAGAGACAAGCCACUCACACCAAACUCUUUGGUGCCUUUGGAAUACGAGCCAGAGGAUAAGCUGUUGGCUGA -----------.((((((((((.((((......))))....))))))))))...(((((....((((((.....))))))(((((((........))))))).........))))).. ( -31.90) >DroAna_CAF1 87345 107 - 1 ------C-----UGGUUAGGUACCCAUCUCGCUAUGGUCUGUACUUGAAGGACAAGCCACUCACCCCCAACGCUCUGGUGCCCCUGGAGUACGAGCCAGAGGACAAGCUUCUGGCGGA ------.-----((((.((((....)))).))))(((..(((.((....)))))..))).......((...((((..(.....)..))))....((((((((.....)))))))))). ( -33.50) >consensus ____________CUUUCAGAUACCCAUCUCGCUAUGGUCUCUACUUGAGAGACAAGCCACUCACACCAAACUCUUUGGUGCCUUUGGAAUACGAGCCAGAGGACAAGCUGCUGGCUGA .................((((....)))).((((..((((((.....)))))).(((......((((((.....))))))(((((((........)))))))....)))..))))... (-23.58 = -24.33 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:09 2006