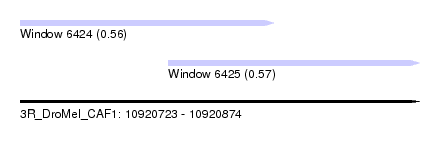
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,920,723 – 10,920,874 |
| Length | 151 |
| Max. P | 0.571561 |

| Location | 10,920,723 – 10,920,819 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10920723 96 + 27905053 UGUCGCCUUUGCUUUGCUUCUUUCUUCGCCUUCGUCAAGAGAAAGAGAGGGCACGAGCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGA .(((((((((..((..((.((((((((((((((.((........))))))))..))).)))))))..)))))).)))))((((.......)))).. ( -31.50) >DroSec_CAF1 191245 96 + 1 UGUCGCCUUUGCUUUGCUUCUUUCUUCGCCUUCGUCAAGAGAAAGAGAGGGCACGAGCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGA .(((((((((..((..((.((((((((((((((.((........))))))))..))).)))))))..)))))).)))))((((.......)))).. ( -31.50) >DroEre_CAF1 190754 90 + 1 UGUUGCUUUUGCUUUGCUUCUUUCUUCGCCUUCGUCAAGAGAAAGAGAGGGCAAGCGCGAGCGAGUGAAGGAGAGCGGCGAGA------UUCUUGA .(((((((((.(((..(((........((((((.((........))))))))..((....)))))..))))))))))))....------....... ( -32.90) >consensus UGUCGCCUUUGCUUUGCUUCUUUCUUCGCCUUCGUCAAGAGAAAGAGAGGGCACGAGCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGA .(((((((((..((..((.((((((((((((((.((........))))))))..))).)))))))..)))))).)))))((((.......)))).. (-27.39 = -27.40 + 0.01)

| Location | 10,920,779 – 10,920,874 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10920779 95 + 27905053 GCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGAGGAUGCUGUU------------GCUGCCUUUUUUGCCUAGCAAUUUAU-UUGUUGUUUGGUUUUUCAA ..(((((((..((((((.(((((((....(((((((....)))).)))))------------)))))))))))..)(.((((((....-..)))))).).)))))).. ( -29.40) >DroSec_CAF1 191301 96 + 1 GCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGAGGAUGCUGUU------------GCUGCCUUUUUUGCCUAGCAAUUUAUUUUGUUGUGUGGUUUUUCGA .((((((((..((((((.(((((((....(((((((....)))).)))))------------)))))))))))..).((((((......)))))).....))))))). ( -30.70) >DroEre_CAF1 190810 98 + 1 GCGAGCGAGUGAAGGAGAGCGGCGAGA------UUCUUGAGGAUGCUGUUGCUGUUGUUGUUGCUGCCUUUUUUCCCUAGCAAUUUAUUUUGAUGUGUGGU----CGA ((.((.(((.(((((..((((((((.(------(((....))))((....)).....)))))))).))))).))).)).))........(((((.....))----))) ( -23.90) >consensus GCGAGAGAGUGAAAGAGAGCGGCGAGAUUCAGAUUCUUGAGGAUGCUGUU____________GCUGCCUUUUUUGCCUAGCAAUUUAUUUUGUUGUGUGGUUUUUCGA ((...((.(..((((((.(((((((((.......)))).((....))...............)))))))))))..))).))........................... (-19.35 = -19.47 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:45 2006