
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,910,843 – 10,910,937 |
| Length | 94 |
| Max. P | 0.829173 |

| Location | 10,910,843 – 10,910,937 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -11.64 |
| Energy contribution | -13.67 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10910843 94 + 27905053 ------GCGGCACAACAACAUAUGUGCAAGUGUGCGCCAGCGAAUUGAACAAUGC------AAAUUACGCAUUAAUU-UUGCAUUUAAUUCGGUGCGAAAGCAAAAG------ ------(((.((((...((....)).....)))))))...((((((((...((((------(((............)-)))))))))))))).(((....)))....------ ( -26.30) >DroPse_CAF1 225614 95 + 1 AAAAGCGCAGCAGAGCAA-----ACGCAAAA-------CGUAAAUUGAACAAUGA------AAAUUACGCAUUUAUUUUUGGCUGCAAUUAAGCGAGAAAGGAAAAAUAGUGG .....(((.((...((..-----..))....-------.((((.((.........------)).)))))).(((.(((((.(((.......))).))))).))).....))). ( -14.00) >DroSec_CAF1 181488 92 + 1 ------GCGGCACAACAACAAAUGUGCAAGU--GCGCCAGCGAAUUGAACAAUGC------AAAUUACGCAUUAAUU-UUGCAUUUAAUUCGGUGCGAAAGCAAAAG------ ------((.(((((........)))))...(--(((((...(((((((...((((------(((............)-)))))))))))))))))))...)).....------ ( -26.20) >DroSim_CAF1 184227 92 + 1 ------GCGGCACAACAACAAAUGUGCAAGU--GCGCCAGCGAAUUGAACAAUGC------AAAUUACGCAUUAAUU-UUGCAUUUAAUUCGGUGCGAAAGCAAAAG------ ------((.(((((........)))))...(--(((((...(((((((...((((------(((............)-)))))))))))))))))))...)).....------ ( -26.20) >DroEre_CAF1 180912 87 + 1 ------GCGGCACAACAA-----GUGCAAGU--GCGCCAGCGAAUUGAACAAUGC------AAAUUACGCAUUAAUU-UCGCAUUUAAUUCGGUGCGAAAGCAAAAG------ ------((.((((.....-----))))...(--(((((...(((((((...((((------((((((.....)))))-).)))))))))))))))))...)).....------ ( -25.00) >DroYak_CAF1 188190 93 + 1 ------GCGGCACAACAA-----GUGCAAGU--GCGCCUGCAAAUUGAACAAUGCGAAUACAAAUUACGCAUUAAUU-UUGCAUUUAAUUCGGUGCGAAAGCAAAAG------ ------((.((((.....-----))))...(--(((((((((((......(((((((((....))).))))))...)-)))))........))))))...)).....------ ( -25.20) >consensus ______GCGGCACAACAA_____GUGCAAGU__GCGCCAGCGAAUUGAACAAUGC______AAAUUACGCAUUAAUU_UUGCAUUUAAUUCGGUGCGAAAGCAAAAG______ ......((.((((..........))))......(((((...((((((((.(((..........)))..(((........))).)))))))))))))....))........... (-11.64 = -13.67 + 2.03)

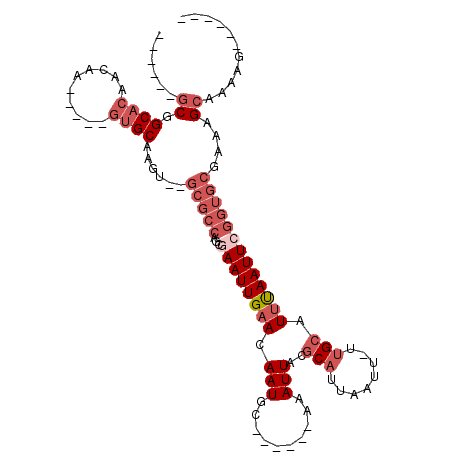
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:38 2006