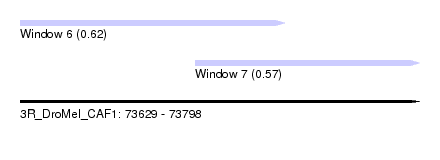
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 73,629 – 73,798 |
| Length | 169 |
| Max. P | 0.618895 |

| Location | 73,629 – 73,741 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.69 |
| Mean single sequence MFE | -42.35 |
| Consensus MFE | -28.69 |
| Energy contribution | -30.08 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 73629 112 + 27905053 CAUUUCGGACGAGGGAGUGGCUCCCAUCGCCAGCGUUUCGAGUGGCUCCCUUAACCUGGGCAGCUUUGUGCCCCAGCACUCCCAAGCUGGCCAAACCAUCAUCCACUCUCAG ......(((.((((((((.((((....((....))....)))).))))))))....(((.((((((.((((....))))....)))))).)))........)))........ ( -44.20) >DroPse_CAF1 17283 106 + 1 UAUCUCCGACGAAGGCGUUGCCCCCAUCACCACCGUUUCCGGUGGCUCGCUCAACCUGGGACACAUUUUGUCCCAGGG---ACAGGC--GCC-AAGCAUCAUCCAGCAGCAG .....((......)).(((((.........(((((....)))))(((((((...(((((((((.....))))))))).---...)))--)..-.)))........))))).. ( -39.10) >DroSec_CAF1 15686 112 + 1 CAUCUCGGACGAGGGAGUGGCUCCCAUCGCCAGCGUUUCGAGUGGCUCCCUUAACCUGGGCAGCUUUGCGCCCCAGCACUCGCAGGCUGGCCAAACCAUCAUCCAGUCUCAG ...((.((((((((((((.((((....((....))....)))).))))))))....(((.((((((.(((..........))))))))).)))............)))).)) ( -43.30) >DroSim_CAF1 19778 112 + 1 CAUCUCGGACGAGGGAGUGGCUCCCAUCGCCAGCGUUUCGAGUGGCUCCCUUAACCUGGGCAGCUUUGCGCCCCAGCACUCGCAGGCUGCCCAAACCAUCAUCCAGUCUCAG ...((.((((((((((((.((((....((....))....)))).))))))))....((((((((((.(((..........)))))))))))))............)))).)) ( -47.90) >DroEre_CAF1 16953 106 + 1 CAUUUCGGACGAGGGAGUGGCCCCCAUUGCCAGCGUCUCGAAUGGCUCCCUCAACCUGGGCAGCUUUGCGCCCCAGCUCUCCCAGGCUGGCCAAACCAUCAUCCAG------ ......(((.((.((..(((((......((((..........))))........((((((.((....((......)).))))))))..)))))..)).)).)))..------ ( -36.50) >DroYak_CAF1 16841 112 + 1 CAUUUCGGACGAGGGAGUGGCCCCCAUCGCUAGCGUUUCGAGUGGCUCCCUCAACCUGGGCAGCUUUGCGUCCCAGCACUCCCAGGGUGGCCAAACCACCAUCCAGCCCCAU ......(((.((((((((.((.(....((....))....).)).))))))))...(((((.((...(((......))))))))))(((((.....))))).)))........ ( -43.10) >consensus CAUCUCGGACGAGGGAGUGGCCCCCAUCGCCAGCGUUUCGAGUGGCUCCCUCAACCUGGGCAGCUUUGCGCCCCAGCACUCCCAGGCUGGCCAAACCAUCAUCCAGUCUCAG ......(((.((((((((.((((....((....))....)))).))))))))....(((.(((((((((......))).....)))))).)))........)))........ (-28.69 = -30.08 + 1.39)

| Location | 73,703 – 73,798 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.26 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 73703 95 + 27905053 AGCACUCCCAAGCUGGCCAAACCAUCAUCCACUCUCAGGCGCCAACCAUUAUCCAGCCACAGGGUCAGCCCAUUGUUCAGCUGCAGUCUAUUAAC ((.(((....(((((..(((.................(((...............)))...(((....))).)))..)))))..)))))...... ( -21.06) >DroSec_CAF1 15760 95 + 1 AGCACUCGCAGGCUGGCCAAACCAUCAUCCAGUCUCAGGCGCCAACCAUUCUCCAGCCACAGGGUCAGCCCAUUGUUCAGCUGCAGUCCAUUAAC .(.(((.((((.(((..(((.................(((...............)))...(((....))).)))..)))))))))).)...... ( -24.26) >DroSim_CAF1 19852 95 + 1 AGCACUCGCAGGCUGCCCAAACCAUCAUCCAGUCUCAGGCGCCAACCAUUCUCCAGCCACAGGGUCAGCCCAUUGUUCAGCUGCAGUCCAUUAAC .(.(((.((((.(((..(((.................(((...............)))...(((....))).)))..)))))))))).)...... ( -22.76) >consensus AGCACUCGCAGGCUGGCCAAACCAUCAUCCAGUCUCAGGCGCCAACCAUUCUCCAGCCACAGGGUCAGCCCAUUGUUCAGCUGCAGUCCAUUAAC .(.(((.(((.((((..(((.................(((...............)))...(((....))).)))..)))))))))).)...... (-21.15 = -21.26 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:51 2006