
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,856,681 – 10,856,820 |
| Length | 139 |
| Max. P | 0.878101 |

| Location | 10,856,681 – 10,856,784 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.85 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
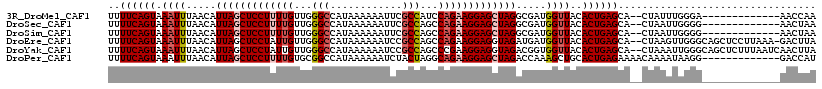
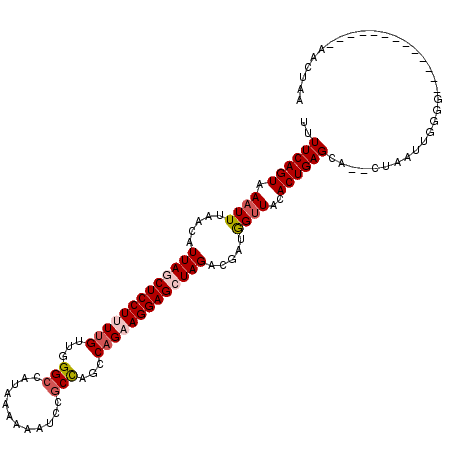
>3R_DroMel_CAF1 10856681 103 - 27905053 UUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGGGCCAUAAAAAAUUCGCCAUCCAGAAGGAGCUAGGCGAUGGUUACACUGAGCA--CUAUUUGGGA-------------AACCAA ..((((((.((((.....(((((((((((((...(((.(........).)))...))))))))))))).....))))..))))))..--....((((..-------------..)))) ( -34.80) >DroSec_CAF1 126034 103 - 1 UUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGGGCCAUAAAAAAUUCGCCAGCCAGAAGGAGCUAGGCGAUGGUUACACUGAGCA--CUAAUUGGGG-------------AACUAA ..((((((.((((.....(((((((((((((...(((.(........).)))...))))))))))))).....))))..))))))..--....((((..-------------..)))) ( -31.10) >DroSim_CAF1 132557 103 - 1 UUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGGGCCAUAAAAAAUUCGCCAGCCAGAAGGAGCUAGGCGAUGGUUACACUGAGCA--CUAAUUGGGG-------------AACUAA ..((((((.((((.....(((((((((((((...(((.(........).)))...))))))))))))).....))))..))))))..--....((((..-------------..)))) ( -31.10) >DroEre_CAF1 128851 115 - 1 UUUUCAGUAAAUUUAACAUUAGCUCCUAUUGUUGGGCCAUAAAAAAUCCGCCAGCCAGAAGGAGGUAGAUGAUGGUUACACUGAGCA--CUAAGUUGGGCAGCUCCUUAAA-GACUUA ..((((((.....(((((.(((...))).)))))((((((.....(((..((..((....)).))..))).))))))..))))))..--.(((((((((.....)))....-)))))) ( -24.50) >DroYak_CAF1 137369 116 - 1 UUUUCAGUAAAUUUAACAUUAGCUCCUAUUGUUGGGCCAUAAAAAAUCCGCCAGCCCGAAGGAGGUAGACGGUGGUUACACUGAGCA--CUAAAUUGGGCAGCUCUUUAAUCAACUUA ....................((((((((...((((((((........(((((.((((....).)))....)))))......)).)).--))))..)))).)))).............. ( -25.44) >DroPer_CAF1 163989 105 - 1 UUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUGCGGCCAUAAAAAAUCUACUAGGCAGAAGGAGCUAGACCAAAGCUGCACUGAGAAAACAAAAUAAGG-------------GACCAU ((((((((..........((((((((((((((..((..............))..))))))))))))))...........))))))))............-------------...... ( -25.64) >consensus UUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGGGCCAUAAAAAAUCCGCCAGCCAGAAGGAGCUAGACGAUGGUUACACUGAGCA__CUAAUUGGGG_____________AACUAA ..((((((.((((.....(((((((((((((...(((............)))...))))))))))))).....))))..))))))................................. (-22.96 = -23.85 + 0.89)

| Location | 10,856,705 – 10,856,820 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.91 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
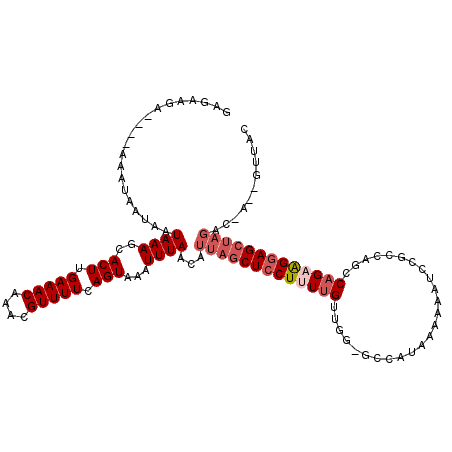
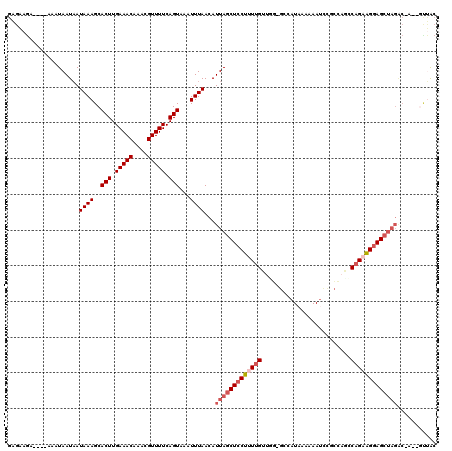
>3R_DroMel_CAF1 10856705 115 - 27905053 GAGAAGA----AAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGG-GCCAUAAAAAAUUCGCCAUCCAGAAGGAGCUAGGCGAUGGUUAC .......----......((((...(((((.(((((....))))).)))..........(((((((((((((...(-((.(........).)))...)))))))))))))))....)))). ( -30.50) >DroPse_CAF1 162827 119 - 1 GAGAAGAAGAAAAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUGCG-GCCAUAAAAAAUCUACUAGGCAGAAGGAGCUAGACCAAAGCUGC .......................((((((.(((((....))))).)))..........((((((((((((((..(-(..............))..))))))))))))))......))).. ( -27.14) >DroWil_CAF1 141527 107 - 1 GUGUAAA----AAAUAAUCAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUUCUUUUGUUUG-CCUAUAAUAAAAACUUCAACCAGCGGCAGCG--------UUGAG .((((..----((((((.....(((((((.(((((....))))).))).(((.....))).))))...)))))).-..)))).........(((((..((....)))--------)))). ( -13.00) >DroYak_CAF1 137406 115 - 1 GAGAAGA----AAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUAUUGUUGG-GCCAUAAAAAAUCCGCCAGCCCGAAGGAGGUAGACGGUGGUUAC .......----......((((...(((((.(((((....))))).)))..........(((.(((((....((((-((................))))))))))).)))...)).)))). ( -21.39) >DroAna_CAF1 130284 109 - 1 GAGAAGA----AAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGUGGCUAU-AAAAAUCUGCCAGGCAGAAGGAGCUAGGC------CAC .......----.............(((((.(((((....))))).)))..........(((((((((((((((.((((...-........)))))))))))))))))))))------... ( -32.60) >DroPer_CAF1 164015 119 - 1 GAGAAGAAGAAAAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUGCG-GCCAUAAAAAAUCUACUAGGCAGAAGGAGCUAGACCAAAGCUGC .......................((((((.(((((....))))).)))..........((((((((((((((..(-(..............))..))))))))))))))......))).. ( -27.14) >consensus GAGAAGA____AAAUAAUAAUAAAGCACUUGAAACAAACGUUUUCAGUAAAUUUAACAUUAGCUCCUUUUGUUGG_GCCAUAAAAAAUCCGCCAGCCAGAAGGAGCUAGAC_A__GUUAC ....................((((..(((.(((((....))))).)))...))))...(((((((((((((.........................)))))))))))))........... (-16.72 = -17.91 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:27 2006