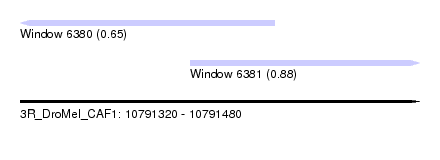
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,791,320 – 10,791,480 |
| Length | 160 |
| Max. P | 0.883086 |

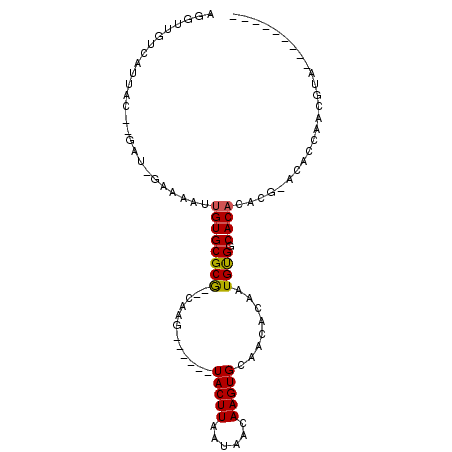
| Location | 10,791,320 – 10,791,422 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -23.14 |
| Energy contribution | -22.64 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10791320 102 - 27905053 -CUUGCGCGCACAAUUUUCAUCGUAAUGACAACCUGCAAGGCAA--AAUGCCUACGGCAAGCGAGAACAUCGCACAUCGCGUGCCAGGGCCGCGACAUAAACUCA -.(((((.((.......((((....))))...(((...((((..--...))))..((((.((((............)))).)))))))))))))).......... ( -29.90) >DroVir_CAF1 87233 88 - 1 -CUCCAGCGCACAAUUUU-GUCGUAAUGACAACCUGCAAGGCA---AAUGCCUAAGGC----GCCAAC-------AUCGCGUGCCUG-GCCUGGGCCGCGACAGC -.....((((......((-(((.....))))).((...((((.---...)))).))))----))....-------.(((((.((((.-....))))))))).... ( -30.60) >DroPse_CAF1 79932 98 - 1 ACUUGCGCGCACAAUUUUCAUCGUAAUGACAACCUGCAGGGCAGAGAAUGCCUACGGCAAGCGCGAACA-------UCGCGUGCCAGGGCCGCGACAUCCACUCA .....(((((.......((((....)))).....(((.(((((.....)))))...))).)))))....-------(((((.((....))))))).......... ( -31.70) >DroEre_CAF1 63062 102 - 1 -CUUGCGCGCACAAUUUUCAUCGUAAUGACAACCUGCAAGGCGA--AAUGCCUACGGCAAGCGAGAACAUCACACAUCGCGUGCCAGGGCCGCGACAUAAACCCA -.(((((.((.......((((....))))...(((...((((..--...))))..((((.((((............)))).)))))))))))))).......... ( -29.20) >DroAna_CAF1 63687 95 - 1 -CUUGUGCGCACAAUUUUCAUCGUAAUGACAACCUGCAAGGCGA--AAUGCCUACGGCAAGCGAGAACA-------UCGCGUGCUAGGGCCGCGACAUCAACUCA -..(((.(((..........((((..((.(.....)))..))))--...((((..((((.((((.....-------)))).)))).)))).))))))........ ( -26.60) >DroPer_CAF1 79480 98 - 1 ACUUGCGCGCACAAUUUUCAUUGUAAUGACAACCUGCAGGGCAGAGAAUGCCUACGGCAAGAGCGAACA-------UCGCGUGCCAGGGCCGCGACAUCCACUCA ..(((((.(((((((....)))))........(((...(((((.....)))))..((((...(((....-------.))).)))))))))))))).......... ( -31.80) >consensus _CUUGCGCGCACAAUUUUCAUCGUAAUGACAACCUGCAAGGCAA__AAUGCCUACGGCAAGCGAGAACA_______UCGCGUGCCAGGGCCGCGACAUCAACUCA ..(((((.((.......((((....))))...(((...(((((.....)))))..((((.(((..............))).)))))))))))))).......... (-23.14 = -22.64 + -0.50)

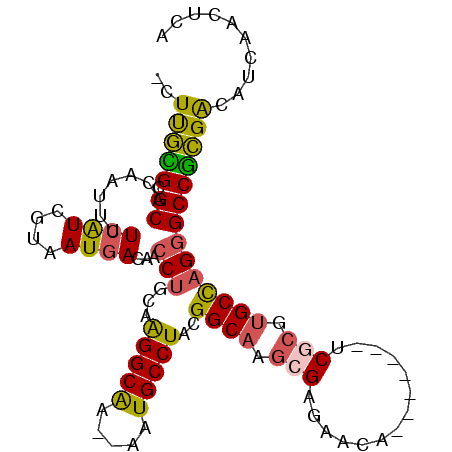
| Location | 10,791,388 – 10,791,480 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -7.38 |
| Energy contribution | -7.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10791388 92 + 27905053 AGGUUGUCAUUAC--GAU-GAAAAUUGUGCGCG--CAAG------UACUUAAUAACAAGUGUCGCACAAUGUGGCACAAACGUACACCAGCGCAAGUUUCUAA ..(((((....))--)))-((((.((((((((.--...)------)............(((((((.....)))))))............)))))).))))... ( -25.50) >DroPse_CAF1 79995 86 + 1 AGGUUGUCAUUAC--GAU-GAAAAUUGUGCGCG--CAAGUACGAGUACUUAAUAACAAGUGCAACACAAUGUGGCACACA---ACAACAACAUA--------- ..((((((((...--.))-))....(((((.((--((.......((((((......)))))).......)))))))))))---)).........--------- ( -19.94) >DroWil_CAF1 70794 97 + 1 AGGUUGUCAUUACACAAUGGAAAAUUGUGCGCACCCAAG------UACUUAAUAACAAGUGCAACACAAUGCGCCACCUACUACCGUGUAUGUGUGUAUAUGU .((.(((.....((((((.....)))))).))).))..(------(((((......))))))...(((((((((.((.(((......))).)))))))).))) ( -22.90) >DroYak_CAF1 64829 92 + 1 AGGUUGUCAUUAC--GAU-GAAAAUUGUGCGCG--CAAG------UACUUAAUAACAAGUGUCGCACAAUGUGGCACAAACGCACACCAACGCAAGUUCCCAA .(((..((((...--.))-)).....((((((.--...)------)............(((((((.....)))))))....)))))))(((....)))..... ( -26.00) >DroAna_CAF1 63748 79 + 1 AGGUUGUCAUUAC--GAU-GAAAAUUGUGCGCA--CAAG------UACUUAAUAACAAGUGCCACACAAUGUGGCACACACGCACACAA-C------------ ..((((((((...--.))-))....((((((..--...(------(........))..(((((((.....)))))))...)))))))))-)------------ ( -23.80) >DroPer_CAF1 79543 86 + 1 AGGUUGUCAUUAC--AAU-GAAAAUUGUGCGCG--CAAGUACGAGUACUUAAUAACAAGUGCAACACAAUGUGGCACACA---ACAACAACAUA--------- ..(((((..((((--(.(-(....((((((...--...))))))((((((......))))))....)).)))))...)))---)).........--------- ( -19.90) >consensus AGGUUGUCAUUAC__GAU_GAAAAUUGUGCGCG__CAAG______UACUUAAUAACAAGUGCAACACAAUGUGGCACACACG_ACACCAACGUA_________ .........................((((((((............(((((......)))))........)))).))))......................... ( -7.38 = -7.18 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:04 2006