
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,775,534 – 10,775,641 |
| Length | 107 |
| Max. P | 0.933773 |

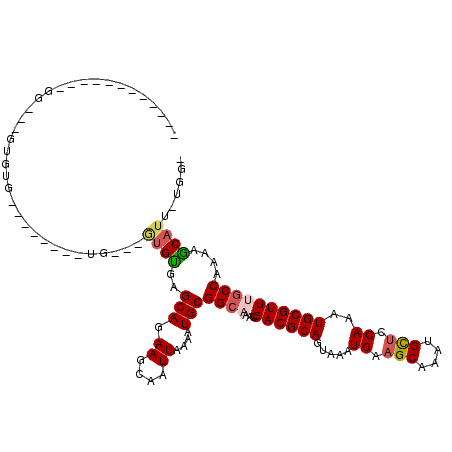
| Location | 10,775,534 – 10,775,627 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10775534 93 + 27905053 ------------GG---UCGUG--------UG---AUGUGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUUGCCAAAAGCAUUCUGG- ------------..---.....--------..---.......(((((((..(((...((((........)))).))).....((((((((.......)))))))).....)).))))).- ( -24.20) >DroVir_CAF1 66581 96 + 1 ------------GG---GUGUG--------UGUCUGUGUAAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCGAAUGUGCCAAAUGCGUUUGCCAAAAGCAUUGUGG- ------------..---...((--------(.(((((....))))).))).....((((((((((((..(((..........))).....((.....)))))))))....)))))....- ( -25.00) >DroGri_CAF1 59699 80 + 1 ----------------------------------UGUGUGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGUUCCAAAUGCGUUUGCCAAAAGC-----GG- ----------------------------------.......((....(((.......)))((((...((((((.....((.(((....))).))..))))))))))....))-----..- ( -18.70) >DroWil_CAF1 53858 90 + 1 ------------AUGCGAUGUU--------U----GUGUGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUGGCCAAACACAU------ ------------..........--------(----((((..(((.((....))....)))(((...(((((((.....((.(((....))).))..))))))))))..))))).------ ( -24.60) >DroMoj_CAF1 67222 116 + 1 GUGUAAGUGUCAGG---GUAUGCGUGCGAUUGUAUGUGCGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGUUCCAAAUGCGUUUGCCAAAAGCAUUGUGG- ((((..(((((...---...(((((..((((((.(.((....)).).))))))...)))))......)))))..........((((((((.......)))))))).....)))).....- ( -28.60) >DroPer_CAF1 61604 96 + 1 ------------AG-AGCUGUG--------UG---AUGUGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUGCCCAAAAGCAUCCUGUG ------------..-.(((..(--------..---(((((((((...((........((((........)))).........))...))))).....))))..).....)))........ ( -22.13) >consensus ____________GG___GUGUG________UG___GUGUGAGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUUGCCAAAAGCAUU_UGG_ ...................................((((........(((.......)))((((...((((((.....((.(((....))).))..))))))))))....))))...... (-16.69 = -17.00 + 0.31)


| Location | 10,775,534 – 10,775,627 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -13.77 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10775534 93 - 27905053 -CCAGAAUGCUUUUGGCAAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCACAU---CA--------CACGA---CC------------ -.(((((.......((((((((((..((((((....)))))).....)))))...)))))(((.......)))))))).......---..--------.....---..------------ ( -25.50) >DroVir_CAF1 66581 96 - 1 -CCACAAUGCUUUUGGCAAACGCAUUUGGCACAUUCGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUUACACAGACA--------CACAC---CC------------ -....(((((....(((((((((.....)).....(((..........)))..)))))))))))).....((..((((......))))..--------))...---..------------ ( -20.20) >DroGri_CAF1 59699 80 - 1 -CC-----GCUUUUGGCAAACGCAUUUGGAACAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCACACA---------------------------------- -..-----((....((((((((((..((((.(....).)))).....)))))...)))))(((.......)))....)).......---------------------------------- ( -16.60) >DroWil_CAF1 53858 90 - 1 ------AUGUGUUUGGCCAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCACAC----A--------AACAUCGCAU------------ ------.(((((..((((.(((((..((((((....)))))).....))))).)...)))(((.......)))........))))----)--------..........------------ ( -23.10) >DroMoj_CAF1 67222 116 - 1 -CCACAAUGCUUUUGGCAAACGCAUUUGGAACAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCGCACAUACAAUCGCACGCAUAC---CCUGACACUUACAC -.....((((....((((((((((..((((.(....).)))).....)))))...)))))(((((.((..(((........)))..)).))).))..))))..---.............. ( -20.40) >DroPer_CAF1 61604 96 - 1 CACAGGAUGCUUUUGGGCAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCACAU---CA--------CACAGCU-CU------------ .....((((....(((((((((((..((((((....)))))).....)))))........(((.......)))...)))))))))---).--------.......-..------------ ( -24.60) >consensus _CCA_AAUGCUUUUGGCAAACGCAUUUGGAACAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCUCACAC___CA________CACAC___CC____________ ........((....((((((((((..((((........)))).....)))))...)))))(((.......)))....))......................................... (-13.77 = -14.30 + 0.53)



| Location | 10,775,548 – 10,775,641 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -25.01 |
| Consensus MFE | -17.76 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10775548 93 + 27905053 AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUUGCCAAAAGCAUUCUGG-----CCG---UGCGC-----AAGU------AU-------- ..(((((((..(((...((((........)))).))).....((((((((.......)))))))).....)).))))).-----..(---(((..-----..))------))-------- ( -28.50) >DroPse_CAF1 62001 101 + 1 AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUGCCCAAAAGCAUCCUGUGGCCCCCU---UGCAC-----AUGUCCAUG----------- .((((..(((.......)))(((...(((((((.....((.(((....))).))..))))))).......((.....)).)))...)---)))..-----.........----------- ( -22.10) >DroGri_CAF1 59705 91 + 1 AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGUUCCAAAUGCGUUUGCCAAAAGC-----GG-----GUAAGCUGUGAUGCAUG------------------- .(((...(((.(((...(((((((...((((((.....((.(((....))).))..))))))))))....))-----).-----.)))..)))..)))...------------------- ( -22.60) >DroMoj_CAF1 67259 102 + 1 AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGUUCCAAAUGCGUUUGCCAAAAGCAUUGUGG-----GUUAACUGCAA-------------GGUUGUGCGGGC .((....((((((....(((((..(((..((((((.......((((((((.......)))))))).......)))))).-----)))..))))).-------------))))))....)) ( -26.84) >DroAna_CAF1 48954 93 + 1 AGCAGAAGCAAUUAAAAUGCGGCAAACGGCGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUUGCCAAAGGCAUUCUGU-----CCC---GGCAC-----UUGU------AU-------- .((((((((..(((...((((.(....).)))).))).....((((((((.......)))))))).....)).))))))-----...---.....-----....------..-------- ( -26.10) >DroPer_CAF1 61620 107 + 1 AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUGCCCAAAAGCAUCCUGUGGCCCCCU---UGCAC-----AUGUCCAUGUAUCUG----- ..((((.(((......(((..((((..((((((.....((.(((....))).))..))))))(((((.........)).)))....)---))).)-----)).....))).))))----- ( -23.90) >consensus AGCAGAAGCAAUUAAAAUGCGGCAAACGACGCAGUAAAUGAAGCAAAUGCUCCAAAUGCGUUUGCCAAAAGCAUUCUGG_____CCU___UGCAC_____AUGU______AU________ .((....(((.......)))((((...((((((.....((.(((....))).))..))))))))))....))................................................ (-17.76 = -18.07 + 0.31)



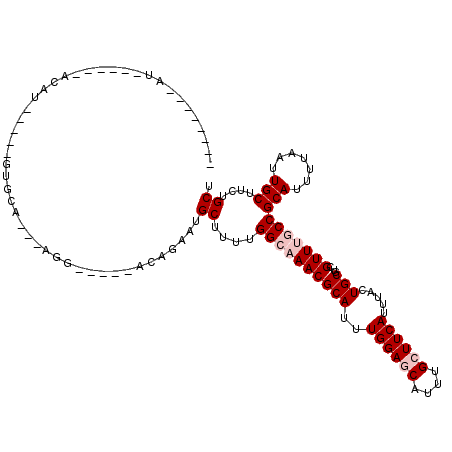
| Location | 10,775,548 – 10,775,641 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.19 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.37 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10775548 93 - 27905053 --------AU------ACUU-----GCGCA---CGG-----CCAGAAUGCUUUUGGCAAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU --------..------....-----..(((---.((-----(.(((((((....((((((((((..((((((....)))))).....)))))...))))))))))))....)))..))). ( -28.70) >DroPse_CAF1 62001 101 - 1 -----------CAUGGACAU-----GUGCA---AGGGGGCCACAGGAUGCUUUUGGGCAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU -----------.........-----..(((---..(((((...(((((((.....(((((((((..((((((....)))))).....)))))....)))))))))))....)))))))). ( -29.00) >DroGri_CAF1 59705 91 - 1 -------------------CAUGCAUCACAGCUUAC-----CC-----GCUUUUGGCAAACGCAUUUGGAACAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU -------------------.((((.....(((....-----..-----)))...((((((((((..((((.(....).)))).....)))))...))))))))).......((....)). ( -19.20) >DroMoj_CAF1 67259 102 - 1 GCCCGCACAACC-------------UUGCAGUUAAC-----CCACAAUGCUUUUGGCAAACGCAUUUGGAACAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU ....(((.....-------------..((((((((.-----....(((((....((((((((((..((((.(....).)))).....)))))...))))))))))))))))))...))). ( -23.20) >DroAna_CAF1 48954 93 - 1 --------AU------ACAA-----GUGCC---GGG-----ACAGAAUGCCUUUGGCAAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGCCGUUUGCCGCAUUUUAAUUGCUUCUGCU --------..------..((-----((((.---(((-----.(.....))))..((((((((((..((((((....)))))).....)))...))))))))))))).....((....)). ( -25.90) >DroPer_CAF1 61620 107 - 1 -----CAGAUACAUGGACAU-----GUGCA---AGGGGGCCACAGGAUGCUUUUGGGCAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU -----((((..((..((.((-----(((((---((.(.(((.(((((...)))))))).(((((..((((((....)))))).....)))))).)))).))))).))...))..)))).. ( -32.50) >consensus ________AU______ACAU_____GUGCA___AGG_____ACAGAAUGCUUUUGGCAAACGCAUUUGGAGCAUUUGCUUCAUUUACUGCGUCGUUUGCCGCAUUUUAAUUGCUUCUGCU ................................................((....((((((((((..((((((....)))))).....)))...)))))))(((.......)))....)). (-17.37 = -18.37 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:02 2006