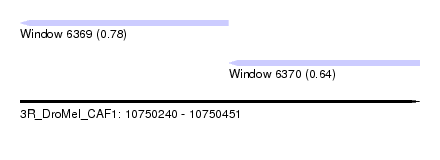
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,750,240 – 10,750,451 |
| Length | 211 |
| Max. P | 0.782471 |

| Location | 10,750,240 – 10,750,350 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10750240 110 - 27905053 AUUAUCCGGGCGCAUUUAUCUCGCAUUGGCAAUUGGAUUUGUAAUGUGACUAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((.(((...((((..((((((((.(((......))))))))))).))))))).(......)....)))))).....(((((((....)))))))........... ( -24.70) >DroPse_CAF1 29255 110 - 1 AUUAUCCGAGCGCAUUUAUCUCGCAUUGGCAAUUGGAUUUGUAAUGUGACCAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((...((..(((.(((.((.((((((((((......)))).)).)))).)).))).))).))...)))))).....(((((((....)))))))........... ( -25.10) >DroEre_CAF1 21906 110 - 1 AUUAUCCGGGCGCAUUUAUCUCGCAUUGGCAAUUGGAUUUGUAAUGUGACUAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((.(((...((((..((((((((.(((......))))))))))).))))))).(......)....)))))).....(((((((....)))))))........... ( -24.70) >DroYak_CAF1 22396 110 - 1 AUUAUCCGGGCGCAUUUAUCUCGCAUUGCCAAUUGGAUUUGUAAUGUGACUAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((.(((...((((..(((((((((.((......))))))))))).))))))).(......)....)))))).....(((((((....)))))))........... ( -24.70) >DroAna_CAF1 20336 110 - 1 AUUAUCCAAACGCAUUUAUCUCACAUUGGCAAUUGGAUUUGUAAUGUGACUAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((....((.((((..((((((((.(((......))))))))))).))))))..(......)....)))))).....(((((((....)))))))........... ( -23.20) >DroPer_CAF1 28470 110 - 1 AUUAUCCGAGCGCAUUUAUCUCGCAUUGGCAAUUGGAUUUGUAAUGUGACCAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((...((..(((.(((.((.((((((((((......)))).)).)))).)).))).))).))...)))))).....(((((((....)))))))........... ( -25.10) >consensus AUUAUCCGAGCGCAUUUAUCUCGCAUUGGCAAUUGGAUUUGUAAUGUGACUAAAGCCAGACAAAACGACAGGAUAACAAAGCAAUGGCAAUGGUCAUUGAAAAGGACAAU .((((((...((..(((.((((((((((.(((......))))))))))).........)).))).))...)))))).....(((((((....)))))))........... (-22.14 = -22.00 + -0.14)

| Location | 10,750,350 – 10,750,451 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -19.78 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10750350 101 - 27905053 CUACGCCCUCAUCCUCAUCCCCCC------GAGCACUUUCCCUCCGGCUGUCACUUAGUACAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU .............(((........------)))............(((.((((((((((.(((......))).)))))))))).((.....))....)))....... ( -21.10) >DroSec_CAF1 21188 99 - 1 CUACGCCCCCAUC--CAACCGCCC------GCGCACUUUCCCUCCGGCUGUCACUUAGUACAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU .............--.....((..------..))...........(((.((((((((((.(((......))).)))))))))).((.....))....)))....... ( -20.70) >DroSim_CAF1 21583 99 - 1 CUACGCCCCCAUU--CAACCCCCC------GCGCACUUUCCCUCCGGCUGUCACUUAGUACAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU ...(((.......--.........------)))............(((.((((((((((.(((......))).)))))))))).((.....))....)))....... ( -20.59) >DroEre_CAF1 22016 104 - 1 CCGCGUAACCACC--CAACGUCCCUCCACAGAGCACU-UCCCUCCGGCUGUCACUUAGAGCAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU ..((((.......--.((((...........(((.(.-.......))))(((((((((.(((......)))...)))))))))))))........))))........ ( -20.19) >DroYak_CAF1 22506 105 - 1 CCCCCCAACCGCC--CAACGUCCCGCCAUCGAGCACUUUCCCUCCGGCUGUCACUUAGUGCAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU .............--.((((....(((...(((........))).))).((((((((((.(((......))).)))))))))))))).................... ( -24.90) >consensus CUACGCCCCCAUC__CAACCCCCC______GAGCACUUUCCCUCCGGCUGUCACUUAGUACAUGUCCUUGUGGACUAAGUGACCGUUUCCCACACAUGCCAGUUCAU .............................................(((.((((((((((.(((......))).)))))))))).((.....))....)))....... (-19.78 = -19.98 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:53 2006