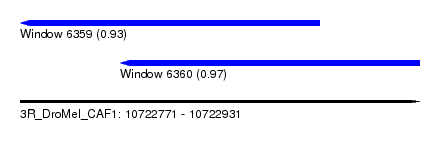
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,722,771 – 10,722,931 |
| Length | 160 |
| Max. P | 0.969583 |

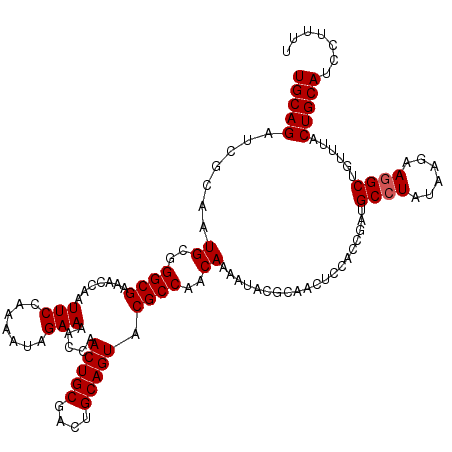
| Location | 10,722,771 – 10,722,891 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -24.60 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10722771 120 - 27905053 UGCAGAUCGCAAUGCCGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACUCCACCGAUGCCUAUAAGAAGGCCGUUUACUGCAUCCUUUU (((((...((...(((((((.......(((.......))).....(((((....))))).))))......(((.(((.(......).))).))).....))).))...)))))....... ( -26.00) >DroSec_CAF1 10505 120 - 1 UGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGAUUGCAGUACGCCAACAAAAUACGCAACUCCACCGAUGCCUAUAAGAAGGCUGUUUACUGCAUCCUUUU (((((...(((.((((((((.......(((.......))).....(((((....))))).)))).........))))...........((((......)))))))...)))))....... ( -27.20) >DroSim_CAF1 10495 120 - 1 UGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGUAACUCCACCGAUGCCUAUAAGAAGGCUGUUUACUGCAUCCUUUU (((((((((...((..((((.......(((.......))).....(((((....))))).))))..))......((......))))))((((......))))......)))))....... ( -27.00) >DroEre_CAF1 11887 120 - 1 UGCAGAUCGAAAUGCGGGCGAAACCAAUUCGAGAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACUCCACCGAUGCCUAUAAGAAGGCUGUUUACUGCAUCCUUUU (((((((((...((((((((.......(((.......))).....(((((....))))).)))).........)))).......))))((((......))))......)))))....... ( -28.40) >DroYak_CAF1 10423 120 - 1 UGCAGAUAGAAAUGCGGGCGAAACCAAUUCCAGAAUUGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACUCCACCGAUGCCUAUAAGAAAGCUGUUUACUGCAUCCUUUU (((((.((((...((.((((....(((((....))))).......(((((....))))).))))......(((.(((.(......).))).)))......))..)))))))))....... ( -28.70) >consensus UGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACUCCACCGAUGCCUAUAAGAAGGCUGUUUACUGCAUCCUUUU (((((.......((..((((.......(((.......))).....(((((....))))).))))..))....................((((......))))......)))))....... (-24.60 = -24.80 + 0.20)

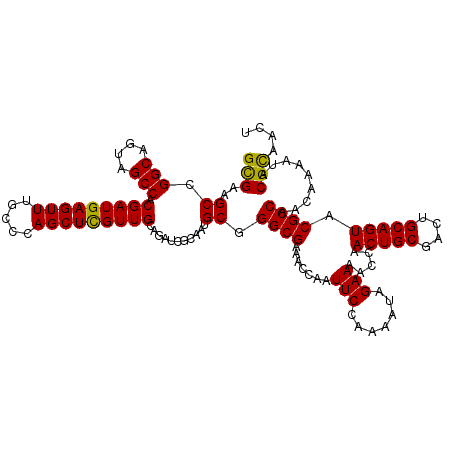
| Location | 10,722,811 – 10,722,931 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -31.10 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10722811 120 - 27905053 GCGAAGCGGGCAGUAGCCACGAUGAGUUUGCCCAGCUCGUUGCAGAUCGCAAUGCCGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACU (((..((((((....))).(((((((((.....))))))))).....)))......((((.......(((.......))).....(((((....))))).)))).........))).... ( -34.70) >DroSec_CAF1 10545 120 - 1 GCGAAGCCGGCAGUAGCCACGAUGAGUUUGCCCAGCUCGUUGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGAUUGCAGUACGCCAACAAAAUACGCAACU (((..((.(((....))).(((((((((.....)))))))))......))..((..((((.......(((.......))).....(((((....))))).))))..)).....))).... ( -33.70) >DroSim_CAF1 10535 120 - 1 GCGAAGCCGGCAGUAGCCACGAUGAGUUUGCCCAGCUCGUUGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGUAACU ((((....(((....))).(((((((((.....)))))))))....))))..((..((((.......(((.......))).....(((((....))))).))))..))............ ( -33.30) >DroEre_CAF1 11927 120 - 1 GCGAAGCGGGCAGUAGCCACGAUGAGUUUGCCCAGCUUGUUGCAGAUCGAAAUGCGGGCGAAACCAAUUCGAGAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACU (((((((((((((...(......)...)))))).))))((((((........))).((((.......(((.......))).....(((((....))))).)))))))......))).... ( -34.50) >DroYak_CAF1 10463 120 - 1 GUGAAGCUGGCAGUAGCCACGAUGAGUUUGCCCAGCUCGUUGCAGAUAGAAAUGCGGGCGAAACCAAUUCCAGAAUUGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACU (((..((((((....))))(((((((((.....)))))))))...........)).((((....(((((....))))).......(((((....))))).)))).........))).... ( -33.40) >consensus GCGAAGCCGGCAGUAGCCACGAUGAGUUUGCCCAGCUCGUUGCAGAUCGCAAUGCGGGCGAAACCAAUUCCAAAAUAGAAAACCAACUGCGACUGCAGUACGCCAACAAAAUACGCAACU (((..((.(((....))).(((((((((.....)))))))))...........)).((((.......(((.......))).....(((((....))))).)))).........))).... (-31.10 = -30.62 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:43 2006