
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,720,988 – 10,721,342 |
| Length | 354 |
| Max. P | 0.997734 |

| Location | 10,720,988 – 10,721,105 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -27.96 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10720988 117 + 27905053 AAGGCAUGGAUGGAAAUAUGUCGGCUUACGGUGGACAACAAGUGGCUGUUUAUUUGCGGCAGACACUUUCGCCGGACUUAUCUGCCAAA---CAAGGGAAACCUUAGAAAGGACGCCAUU ..((((((........))))))(((...(((((((.....(((((((((......)))))...)))))))))))..(((.((((.....---..(((....))))))).)))..)))... ( -35.91) >DroSec_CAF1 8736 120 + 1 AAGGAAUGGAUGGAAAUAUGUCGGCUUACGUUGGACAACAAGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUU ....(((((.(((..(((.((((((....(((....)))((((((((((......)))))...)))))..))).))).)))...)))......((((....))))..........))))) ( -32.30) >DroSim_CAF1 8723 120 + 1 AAGGAAUGGAUGGAAAUAUGUCGGCUUACGUUGGACAACAAGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUU ....(((((.(((..(((.((((((....(((....)))((((((((((......)))))...)))))..))).))).)))...)))......((((....))))..........))))) ( -32.30) >DroEre_CAF1 10117 120 + 1 AAGGUAUGGAUGGAAACAUGUCGUCUCACGUUGGACAACAAGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCGGACUUAUCUGCCAAAUAGGAAGGGAAACCUUAAAAAGGACGCCAUU .........(((....)))((.((((......)))).)).((((((.((((......((((((.....((....))....)))))).......((((....)))).....)))))))))) ( -35.30) >DroYak_CAF1 8651 120 + 1 AAGGAAUGGAUGGAAAUAUGUCGUCUUACGUUGGACAACAAGUGGCUGUUCAUUUGCGGUCGACACUUUCGUCAGACUUAUCUGCCAAAUAGCAAGGGAAACCUUAGAAAGGACGCCAUU ...((((((((((..((.(((.((((......)))).))).))..))))))))))..((.((...((((((.((((....)))).).......((((....)))).)))))..))))... ( -36.30) >consensus AAGGAAUGGAUGGAAAUAUGUCGGCUUACGUUGGACAACAAGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAGGACGCCAUU ...((((((((((..((.(((.(.((......)).).))).))..))))))))))((((((((.....((....))....)))))).......((((....)))).........)).... (-27.96 = -28.08 + 0.12)

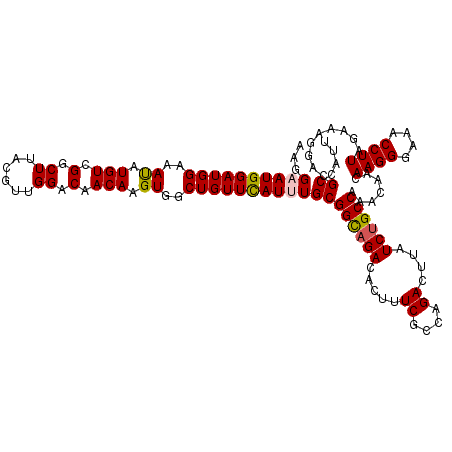

| Location | 10,721,028 – 10,721,145 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -39.04 |
| Consensus MFE | -37.52 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721028 117 + 27905053 AGUGGCUGUUUAUUUGCGGCAGACACUUUCGCCGGACUUAUCUGCCAAA---CAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCG ((((((.(((..(((..((((((.....((....))....))))))...---.((((....)))).)))..))))))))).....(((((.....))))).((((.((.....)))))). ( -41.00) >DroSec_CAF1 8776 120 + 1 AGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCG ((((((.(((..(((..((((((.....((....))....)))))).......((((....)))).)))..))))))))).....(((((.....))))).((((.((.....)))))). ( -40.30) >DroSim_CAF1 8763 120 + 1 AGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCG ((((((.(((..(((..((((((.....((....))....)))))).......((((....)))).)))..))))))))).....(((((.....))))).((((.((.....)))))). ( -40.30) >DroEre_CAF1 10157 120 + 1 AGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCGGACUUAUCUGCCAAAUAGGAAGGGAAACCUUAAAAAGGACGCCAUUCCAGAAUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUUG ((((((.((((......((((((.....((....))....)))))).......((((....)))).....))))))))))......((((.....))))..((((.((.....)))))). ( -36.80) >DroYak_CAF1 8691 120 + 1 AGUGGCUGUUCAUUUGCGGUCGACACUUUCGUCAGACUUAUCUGCCAAAUAGCAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAAGAGGAGGAUCCCCUUG .(((((.((((......(((((((......))).))))..(((((......))((((....)))))))..)))))))))(((...(((((.....)))))))).((((.(....))))). ( -36.80) >consensus AGUGGCUGUUCAUUUGCGGCAGACACUUUCGCCAGACUUAUCUGCCAAACAACAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCG .(((((.((((.(((..((((((.....((....))....)))))).......((((....)))).))).)))))))))(((...(((((.....)))))...))).((((....)))). (-37.52 = -37.88 + 0.36)


| Location | 10,721,028 – 10,721,145 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.46 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721028 117 - 27905053 CGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUG---UUUGGCAGAUAAGUCCGGCGAAAGUGUCUGCCGCAAAUAAACAGCCACU .((((((((((...(((...(((((.....)))))...)))..)).))))))))...((((....(((---(((((((((((..((....))...))))))))..))))))..))))... ( -40.10) >DroSec_CAF1 8776 120 - 1 CGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCUUUUCUAAGGUUUCCCUUGUUGUUUGGCAGAUAAGUCUGGCGAAAGUGUCUGCCGCAAAUGAACAGCCACU .((((((......)))))).(((((.....))))).......((((.((.....(((((....)))))((((..((((((((..((....))...))))))))))))..))...)))).. ( -40.20) >DroSim_CAF1 8763 120 - 1 CGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCUUUUCUAAGGUUUCCCUUGUUGUUUGGCAGAUAAGUCUGGCGAAAGUGUCUGCCGCAAAUGAACAGCCACU .((((((......)))))).(((((.....))))).......((((.((.....(((((....)))))((((..((((((((..((....))...))))))))))))..))...)))).. ( -40.20) >DroEre_CAF1 10157 120 - 1 CAAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAAUUCUGGAAUGGCGUCCUUUUUAAGGUUUCCCUUCCUAUUUGGCAGAUAAGUCCGGCGAAAGUGUCUGCCGCAAAUGAACAGCCACU ...(((((......))).)).......(((((..(((.((((.((...(((.....)))...)).)))).....((((((((..((....))...))))))))......)))...))))) ( -35.20) >DroYak_CAF1 8691 120 - 1 CAAGGGGAUCCUCCUCUUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUGCUAUUUGGCAGAUAAGUCUGACGAAAGUGUCGACCGCAAAUGAACAGCCACU ((((((((.(((........(((((.....)))))..(((((.((...)).)))))))).)))))))).....(((((((....))).((....))..................)))).. ( -33.30) >consensus CGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUGCUAUUUGGCAGAUAAGUCUGGCGAAAGUGUCUGCCGCAAAUGAACAGCCACU ((((((((.(((........(((((.....)))))..(((((.((...)).)))))))).))))))))......((((((((..((....))...))))))))................. (-34.66 = -34.46 + -0.20)

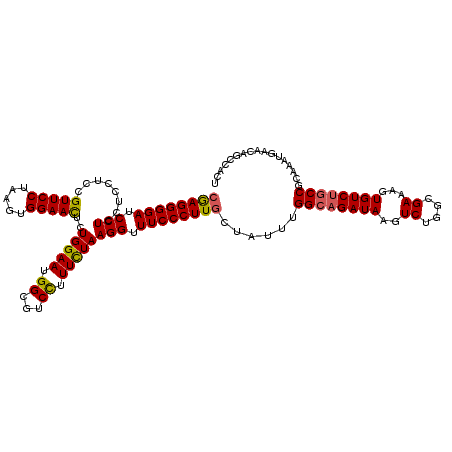

| Location | 10,721,068 – 10,721,185 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721068 117 + 27905053 UCUGCCAAA---CAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCGGAAAGAGCAUCAAAAGUAUGAAAUCAUGUGCUAAUAGAAG ((((.....---.((((....))))......((.(((.((((...(((((.....))))).((((.((.....)))))))))).).)).))...(((((........)))))..)))).. ( -34.50) >DroSec_CAF1 8816 120 + 1 UCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCGGAAAGACCGUCAAAAGUAUGAAAUCAUGUGCUAAUAGAAG ((((.........((((....))))......((((.(.((((...(((((.....))))).((((.((.....)))))))))).)..))))...(((((........)))))..)))).. ( -35.30) >DroSim_CAF1 8803 120 + 1 UCUGCCAAACAACAAGGGAAACCUUAGAAAAGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCGGAAAGACCGUCAAAAGUAUGAAAUCAUGUGCUAAUAGAAG ((((.........((((....))))......((((.(.((((...(((((.....))))).((((.((.....)))))))))).)..))))...(((((........)))))..)))).. ( -35.30) >DroEre_CAF1 10197 120 + 1 UCUGCCAAAUAGGAAGGGAAACCUUAAAAAGGACGCCAUUCCAGAAUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUUGGAAAGUCCAUUAAAACUAUGAAAUCAUGUUCUAAUAGAAG ....((.....))((((....)))).....((((....((((((..((((.....)))).((.(((......))))))))))).))))((((.(((.(((....)))))).))))..... ( -30.90) >DroYak_CAF1 8731 120 + 1 UCUGCCAAAUAGCAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAAGAGGAGGAUCCCCUUGGAAAGAGCAUCACAAGCAUGAAAUCAUGUUCUAAUAAAAG ..(((......)))(((....)))((((...((.(((.((((...(((((.....)))))....((((.(....))))))))).).)).))....(((((....)))))))))....... ( -32.10) >consensus UCUGCCAAACAACAAGGGAAACCUUAGAAAGGACGCCAUUCCAGAGUUCCACUUAGGAACGGAGGAGGAGGAUCCCCUCGGAAAGACCAUCAAAAGUAUGAAAUCAUGUGCUAAUAGAAG .............((((....)))).............((((...(((((.....))))).((((.((.....)))))))))).............((((....))))............ (-24.92 = -24.92 + -0.00)

| Location | 10,721,068 – 10,721,185 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -23.74 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721068 117 - 27905053 CUUCUAUUAGCACAUGAUUUCAUACUUUUGAUGCUCUUUCCGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUG---UUUGGCAGA .........((.((...............((((((..((((((((((......)))))).(((((.....)))))...)))).)))))).....(((((....)))))---..))))... ( -33.20) >DroSec_CAF1 8816 120 - 1 CUUCUAUUAGCACAUGAUUUCAUACUUUUGACGGUCUUUCCGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCUUUUCUAAGGUUUCCCUUGUUGUUUGGCAGA ((.(((.(((((...((..((.((.....((((.((.((((((((((......)))))).(((((.....)))))...)))).)))))).....)).))..))...)))))..))).)). ( -32.10) >DroSim_CAF1 8803 120 - 1 CUUCUAUUAGCACAUGAUUUCAUACUUUUGACGGUCUUUCCGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCUUUUCUAAGGUUUCCCUUGUUGUUUGGCAGA ((.(((.(((((...((..((.((.....((((.((.((((((((((......)))))).(((((.....)))))...)))).)))))).....)).))..))...)))))..))).)). ( -32.10) >DroEre_CAF1 10197 120 - 1 CUUCUAUUAGAACAUGAUUUCAUAGUUUUAAUGGACUUUCCAAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAAUUCUGGAAUGGCGUCCUUUUUAAGGUUUCCCUUCCUAUUUGGCAGA ..((((((((((((((....))).)))))))))))(((...((((((((((...(((...(((((.....)))))...)))..)).)))))))).)))......((.((.....)).)). ( -33.50) >DroYak_CAF1 8731 120 - 1 CUUUUAUUAGAACAUGAUUUCAUGCUUGUGAUGCUCUUUCCAAGGGGAUCCUCCUCUUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUGCUAUUUGGCAGA ......((((((((((....)))).....((((((..((((((((((.....))))))..(((((.....)))))...)))).))))))..))))))((....))(((((....))))). ( -35.50) >consensus CUUCUAUUAGCACAUGAUUUCAUACUUUUGAUGGUCUUUCCGAGGGGAUCCUCCUCCUCCGUUCCUAAGUGGAACUCUGGAAUGGCGUCCUUUCUAAGGUUUCCCUUGCUAUUUGGCAGA ((.(((.......(((....))).................((((((((.(((........(((((.....)))))..(((((.((...)).)))))))).)))))))).....))).)). (-23.74 = -22.98 + -0.76)



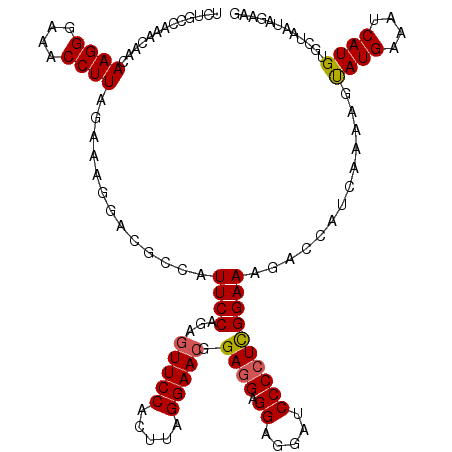
| Location | 10,721,185 – 10,721,305 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721185 120 + 27905053 UAAUAUCAGUUUCAAGAGCGAGGAUUCUGGCAUUGACGAGGAAAGCUUUAAGGAGAGUAAGCCUCAUAAUAAUCCAUGCGCCAAUUGCCUUGAUCUCCAACAGCAAUUGGCAGCAGCUCA ...............((((..(((((..(.......)((((...(((((....)))))...)))).....))))).((((((((((((.(((.....)))..))))))))).))))))). ( -37.60) >DroSec_CAF1 8936 117 + 1 UAAUAUCAGUUUUAAAAGCGAGGAUUCUGGCAUCGACGAGGAAAGCUUCAAGGAGAGUAAGCCUCAU---AAUCCAUGCGCCAAUUGCCUUGAUCUUCAACAGCAAUUGGCAGCUGCUCA ......(((((......(((.(((((..(((.((..(((((....))))..)..))....)))....---))))).)))(((((((((.(((.....)))..)))))))))))))).... ( -35.00) >DroSim_CAF1 8923 117 + 1 UAAUAUCAGUUUCAAAAGCGAGGAUUCUGGCAUCGACGAGGAAAGCUUCAGGGAGAGUAAGCCUCAU---AAUCCAUGCGCCAAUUGCCUUGAUCUUCAACAGCAAUUGGCAGCUGCUCA ......(((((......(((.(((((..(((.((..(((((....))))..)..))....)))....---))))).)))(((((((((.(((.....)))..)))))))))))))).... ( -35.00) >DroEre_CAF1 10317 117 + 1 CAAUAUAAGUUUCAAGAGCGAAGAUUCUGGCAUCGACGAGGAAAGCUUCAAGGAGAGAAAUCCCCAU---GAUCCAUGUGCCAAUUGCCUUGAUCUUAAACAGCAAUUGGCAGCUGCUCU ..............((((((.......(((.((((..((((....))))..((.((....)).)).)---))))))..((((((((((..((........))))))))))))..)))))) ( -33.80) >DroYak_CAF1 8851 117 + 1 CAUUAUAAGUUUCAAGAGCGACGAUUCUGGCAUCGACGAGGAAAGUUUCAAGGAGAGAAAGCCACAC---GAUCCAUGCGCCAAUUGUCUUGAUCUUAAACAGCAAUUAGCAGCUGCUCU ..............((((((((((((...((((.((((.((....((((.....))))...))...)---).)).))))...)))))))............(((........)))))))) ( -25.20) >consensus UAAUAUCAGUUUCAAGAGCGAGGAUUCUGGCAUCGACGAGGAAAGCUUCAAGGAGAGUAAGCCUCAU___AAUCCAUGCGCCAAUUGCCUUGAUCUUCAACAGCAAUUGGCAGCUGCUCA ...............((((..(((((..(((.((..(((((....))))..)..))....))).......)))))..(((((((((((..((........))))))))))).)).)))). (-24.40 = -24.56 + 0.16)



| Location | 10,721,185 – 10,721,305 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.85 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721185 120 - 27905053 UGAGCUGCUGCCAAUUGCUGUUGGAGAUCAAGGCAAUUGGCGCAUGGAUUAUUAUGAGGCUUACUCUCCUUAAAGCUUUCCUCGUCAAUGCCAGAAUCCUCGCUCUUGAAACUGAUAUUA .(((((((.((((((((((.(((.....)))))))))))))))).((((((((((((((....((........))....)))))).)))).....))))..))))............... ( -33.50) >DroSec_CAF1 8936 117 - 1 UGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU---AUGAGGCUUACUCUCCUUGAAGCUUUCCUCGUCGAUGCCAGAAUCCUCGCUUUUAAAACUGAUAUUA .((((....((((((((((.(((.....)))))))))))))((((.(((.---..(((((((.(.......))))))))....))).))))..........))))............... ( -34.80) >DroSim_CAF1 8923 117 - 1 UGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU---AUGAGGCUUACUCUCCCUGAAGCUUUCCUCGUCGAUGCCAGAAUCCUCGCUUUUGAAACUGAUAUUA .((((....((((((((((.(((.....)))))))))))))((((.(((.---..(((((((.(.......))))))))....))).))))..........))))............... ( -34.80) >DroEre_CAF1 10317 117 - 1 AGAGCAGCUGCCAAUUGCUGUUUAAGAUCAAGGCAAUUGGCACAUGGAUC---AUGGGGAUUUCUCUCCUUGAAGCUUUCCUCGUCGAUGCCAGAAUCUUCGCUCUUGAAACUUAUAUUG (((((.(.(((((((((((............)))))))))))).((((((---(((((((((((.......))))...))))))).))).)))........))))).............. ( -37.10) >DroYak_CAF1 8851 117 - 1 AGAGCAGCUGCUAAUUGCUGUUUAAGAUCAAGACAAUUGGCGCAUGGAUC---GUGUGGCUUUCUCUCCUUGAAACUUUCCUCGUCGAUGCCAGAAUCGUCGCUCUUGAAACUUAUAAUG .(((((((........)))))))....((((((....(((((..((((((---(((.((.((((.......))))....)).)).)))).)))....))))).))))))........... ( -29.80) >consensus UGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU___AUGAGGCUUACUCUCCUUGAAGCUUUCCUCGUCGAUGCCAGAAUCCUCGCUCUUGAAACUGAUAUUA .((((.((.((((((((((............)))))))))))).((((((...((((((.....((.....))......)))))).))).)))........))))............... (-27.78 = -27.50 + -0.28)



| Location | 10,721,225 – 10,721,342 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -34.24 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.50 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.536016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10721225 117 - 27905053 CUCUUGC---CUUCUUUCCUCAAGUUGCUGAAUGCGAAGAUGAGCUGCUGCCAAUUGCUGUUGGAGAUCAAGGCAAUUGGCGCAUGGAUUAUUAUGAGGCUUACUCUCCUUAAAGCUUUC .....((---(((.....((((..((((.....))))...)))).(((.((((((((((.(((.....))))))))))))))))...........))))).................... ( -32.70) >DroSec_CAF1 8976 114 - 1 CUCUCGC---CUUUUUUCCUCCAGCUGCUGAAUGCGAAAAUGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU---AUGAGGCUUACUCUCCUUGAAGCUUUC .....((---(((...(((...(((((((..((......)).)))))))((((((((((.(((.....)))))))))))))....)))..---..))))).................... ( -39.00) >DroSim_CAF1 8963 114 - 1 CUCUCGC---CUUCUUUCCUCCAGCUGCUGAAUGCGAAAAUGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU---AUGAGGCUUACUCUCCCUGAAGCUUUC .....((---(((...(((...(((((((..((......)).)))))))((((((((((.(((.....)))))))))))))....)))..---..))))).................... ( -38.30) >DroEre_CAF1 10357 114 - 1 CUCUUGC---UUUUUUUCCUCCACCUGCUGAAUGCGAAGAAGAGCAGCUGCCAAUUGCUGUUUAAGAUCAAGGCAAUUGGCACAUGGAUC---AUGGGGAUUUCUCUCCUUGAAGCUUUC ....(((---((((((((........((.....)))))))))))))(((((((((((((............)))))))))).......((---(.(((((....))))).)))))).... ( -37.10) >DroYak_CAF1 8891 117 - 1 CUCUUGUUCUUUUUUUUCCUCCAACUGCUGAACGCGAAGAAGAGCAGCUGCUAAUUGCUGUUUAAGAUCAAGACAAUUGGCGCAUGGAUC---GUGUGGCUUUCUCUCCUUGAAACUUUC ....(((((((((((....((........))....)))))))))))(((((((((((((...........)).))))))))(((((...)---)))))))((((.......))))..... ( -24.10) >consensus CUCUUGC___CUUUUUUCCUCCAGCUGCUGAAUGCGAAGAUGAGCAGCUGCCAAUUGCUGUUGAAGAUCAAGGCAAUUGGCGCAUGGAUU___AUGAGGCUUACUCUCCUUGAAGCUUUC .....((.........(((...(((((((.............)))))))((((((((((............))))))))))....))).......(((.....)))........)).... (-23.26 = -23.50 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:39 2006