| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,705,097 – 10,705,270 |
| Length | 173 |
| Max. P | 0.958461 |

| Location | 10,705,097 – 10,705,196 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -15.30 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10705097 99 - 27905053 CAACAAAAGGGAA-AUCGUGAUGUAAACAAAAAGCAGCUGACUUCAGCUGUUGCAGCCCUCGUUGGCCUGCGUGAAACGAUCUGGCAACGUCAUGCAUAA .............-..((((((((...((...((((((((....))))))))((((.((.....)).))))...........))...))))))))..... ( -29.60) >DroSim_CAF1 146 79 - 1 CAACAAAAGGGAU-A-CGCGAUGUAAACAAAAGACAGCUGACUUCAGCUGUUGCAGCCCU-------------------AUCUGGCAACGCCGGGCAUAA .......((((..-.-.((..((....))...((((((((....))))))))))..))))-------------------.((((((...))))))..... ( -25.40) >DroYak_CAF1 146 100 - 1 CAACAAGAGAGAGGAACGCGAUGUAAACAAAAAACAGCUGACUUAAGCUGUUGCAGCCCUAUUGGAAGUGUGUGAGUAGAUCUGGCAACGCCGAGAAUAC ...(((...((.((...((..((....))...(((((((......)))))))))..)))).)))...((((((.((.....)).)).))))......... ( -21.70) >consensus CAACAAAAGGGAA_A_CGCGAUGUAAACAAAAAACAGCUGACUUCAGCUGUUGCAGCCCU__U_G___UG_GUGA___GAUCUGGCAACGCCGAGCAUAA .......((((......((..((....))...((((((((....))))))))))..))))......................((((...))))....... (-15.30 = -14.87 + -0.43)

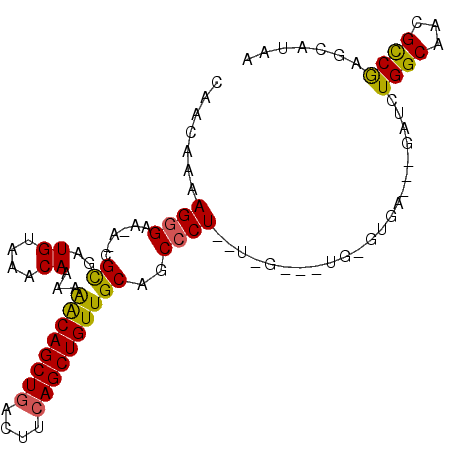
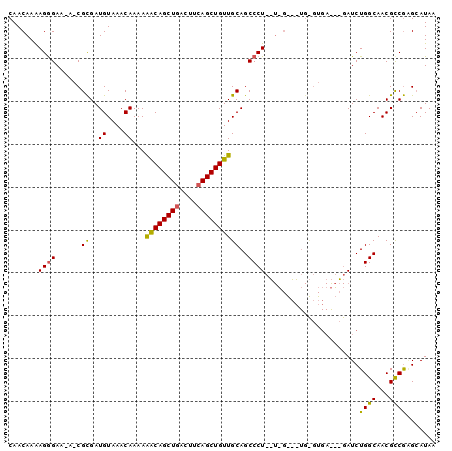
| Location | 10,705,117 – 10,705,235 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.89 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -23.51 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10705117 118 - 27905053 UG-GGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAA-AUCGUGAUGUAAACAAAAAGCAGCUGACUUCAGCUGUUGCAGCCCUCGUUGGCCUGCGUGAAACGA .(-((((((((....))))))).)).(((((((..(((((........(((..-...((.......))....((((((((....))))))))....))))))))..)))))))....... ( -41.60) >DroSim_CAF1 166 99 - 1 UGAGGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAU-A-CGCGAUGUAAACAAAAGACAGCUGACUUCAGCUGUUGCAGCCCU-------------------A .(..(((((((....)))))))..)(((((...................(..(-(-(.....)))..)....((((((((....)))))))))).)))..-------------------. ( -27.10) >DroYak_CAF1 166 119 - 1 UG-GGGAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAGAGAGAGGAACGCGAUGUAAACAAAAAACAGCUGACUUAAGCUGUUGCAGCCCUAUUGGAAGUGUGUGAGUAGA ((-(((...(((((((((((((.((......))....((....)).....))))))))).))))........(((((((......)))))))....)))))................... ( -30.50) >consensus UG_GGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAA_A_CGCGAUGUAAACAAAAAACAGCUGACUUCAGCUGUUGCAGCCCU__U_G___UG_GUGA___GA ....(((((((....)))))))...(((((............(((...(.(.....).)..)))........((((((((....)))))))))).)))...................... (-23.51 = -22.97 + -0.55)

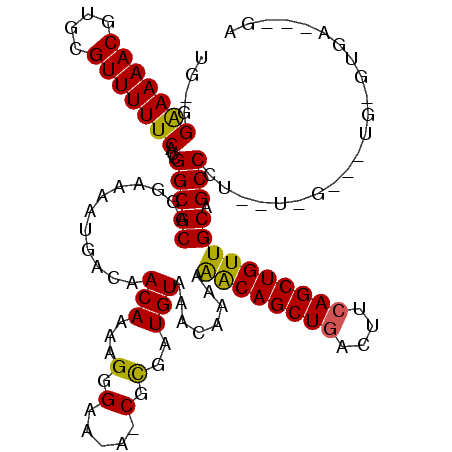
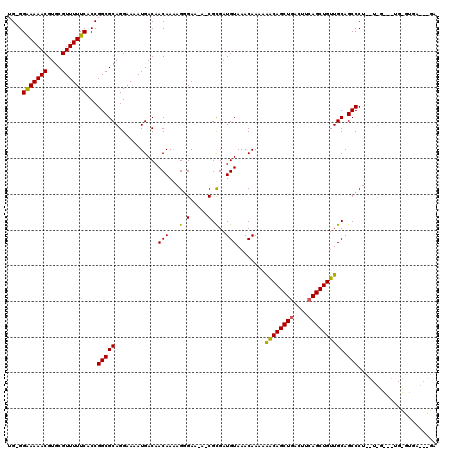
| Location | 10,705,157 – 10,705,270 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10705157 113 - 27905053 CGUGUAUAAAUAGCUUUUGUUUAUGUCGGUGAAAAUG-GGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAA-AUCGUGAUGUAAACAAAAAGCAGCUG ............(((((((((((((((.(((....((-((((((((....))))))).))).)))......((((...(.....)...-.))))))))))))).))))))..... ( -27.50) >DroSim_CAF1 187 113 - 1 CGUUUAUAAAUAGCUUUUGUUUAUGUCGGUGAAAAUGAGGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAU-A-CGCGAUGUAAACAAAAGACAGCUG ............(((((((((((((((((((........(((((((....))))))).((......))...................)-)-).))))))))))))))).)..... ( -27.70) >DroYak_CAF1 206 114 - 1 CGUGUAUAAAUAGCUUUUGUUUAUGUCGGUGAAAAUG-GGGAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAGAGAGAGGAACGCGAUGUAAACAAAAAACAGCUG ..........(((((((((((((((((((((....((-((((((((....))))))).))).))).(........).................)))))))))))))....))))) ( -28.00) >consensus CGUGUAUAAAUAGCUUUUGUUUAUGUCGGUGAAAAUG_GGAAAAACGUGCGUUUUUCACCGGCGCAGGAAAAUGACAACAAAAGGGAA_A_CGCGAUGUAAACAAAAAACAGCUG ..........((((((((((((((((((...........(((((((....))))))).((......)).........................)))))))))))))....))))) (-24.81 = -24.37 + -0.44)

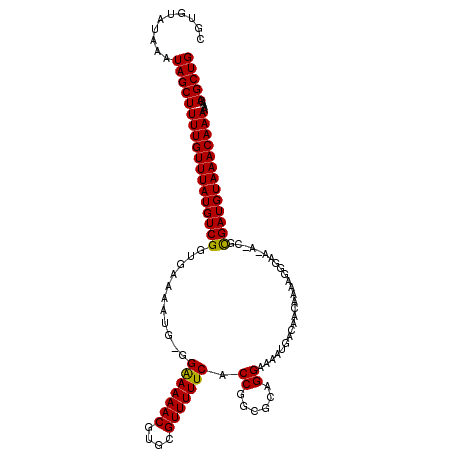
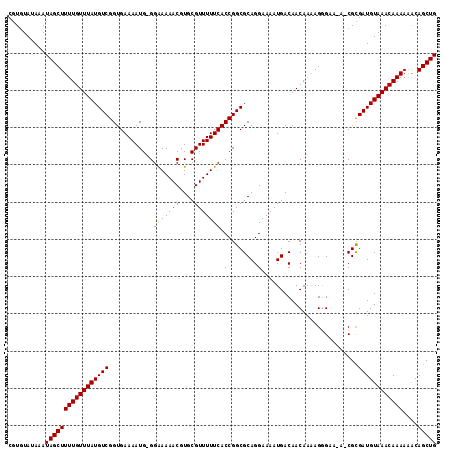
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:27 2006