
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,700,223 – 10,700,324 |
| Length | 101 |
| Max. P | 0.902533 |

| Location | 10,700,223 – 10,700,324 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -11.86 |
| Energy contribution | -13.37 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10700223 101 + 27905053 GAUAGGUGCAGAUGUUUUGUGGCGCUGUGCAUGCAAAUCGUGGAGCAGCACAGUGAAAAA-AAUGUCUAAAAUGGUA--A-UUUACUAAUAUGUAUAAUACGGAU .....(((((((((((((....((((((((.(((..........)))))))))))...))-)))))))....(((((--.-..)))))....))))......... ( -27.20) >DroSec_CAF1 41530 103 + 1 GAUAGGUGCAGAUGUUUUGUGGCGCUGUGCAUGCAAAUGGUGGAGCGGCACAGUAAAAAAUAAUGUCUACAAUGGUA--ACAUUACUAAUAUAUAGAAUACUAAU .........((.(((((((((..(((((((.(((..........))))))))))......((((((.(((....)))--))))))......)))))))))))... ( -22.90) >DroSim_CAF1 44736 103 + 1 GAUAGGUGCAGAUGUUUUGUGGCGCUGUGAAUGCAAAUGGUGGAGCGGCACAGUAAAAAAUAAUGUCUACAAUGGUA--ACUUUACUAAUAUAUAUAAUACUAAU ..(((.((.((((((((((((.((((.((.((....))..)).)))).)))))........))))))).)).(((((--....)))))............))).. ( -18.90) >DroEre_CAF1 38809 88 + 1 GAUAGGUGCAGAUGUUUUGUGGUGCUGUGCAUGCAAAUGGUGGAGCAGCACAGUGAAAAAUAAUGUGUGCAGUGGGA-----------UACCAUCUG------AU ..(((.((((.(((((...(..((((((((.(((..........)))))))))))..)...))))).))))((((..-----------..)))))))------.. ( -26.70) >DroYak_CAF1 34018 99 + 1 GAUAGGUGCAGAUGUUCUGUGGCGCUGUGCAUGCAAAUGGUGGAGCAGCACAGUGAAAAAUAAUGUUUGCCAUAGGAAAACAUUACUACACCAUGUA------AC ....((((.(((((((((((((((((((((.(((..........))))))))))...((((...))))))))))))...))))..)).)))).....------.. ( -31.50) >DroAna_CAF1 17752 82 + 1 GAUAGGUGCAGAUGGUUUUUCGCACAAAGCAUGCAAAUGG--GAGAAGUACAGUGAAAAAA--UUUGAG--AAGGUA-----------GUCCAUCCA------AG ..........(((((((((((((......(((....)))(--........).))))))))(--(((...--.)))).-----------..)))))..------.. ( -11.50) >consensus GAUAGGUGCAGAUGUUUUGUGGCGCUGUGCAUGCAAAUGGUGGAGCAGCACAGUGAAAAAUAAUGUCUACAAUGGUA__A__UUACUAAUACAUAUA______AU .........(((((((......((((((((.(((..........)))))))))))......)))))))..................................... (-11.86 = -13.37 + 1.50)

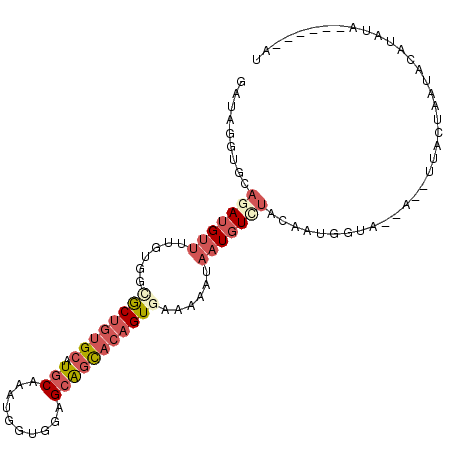

| Location | 10,700,223 – 10,700,324 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -18.19 |
| Consensus MFE | -6.24 |
| Energy contribution | -7.27 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10700223 101 - 27905053 AUCCGUAUUAUACAUAUUAGUAAA-U--UACCAUUUUAGACAUU-UUUUUCACUGUGCUGCUCCACGAUUUGCAUGCACAGCGCCACAAAACAUCUGCACCUAUC ....(((...(((......)))..-.--))).....((((..((-((...(.(((((((((..........))).)))))).)....))))..))))........ ( -15.80) >DroSec_CAF1 41530 103 - 1 AUUAGUAUUCUAUAUAUUAGUAAUGU--UACCAUUGUAGACAUUAUUUUUUACUGUGCCGCUCCACCAUUUGCAUGCACAGCGCCACAAAACAUCUGCACCUAUC ((((((((.....)))))))).....--......((((((............((((((.((..........))..))))))............))))))...... ( -17.85) >DroSim_CAF1 44736 103 - 1 AUUAGUAUUAUAUAUAUUAGUAAAGU--UACCAUUGUAGACAUUAUUUUUUACUGUGCCGCUCCACCAUUUGCAUUCACAGCGCCACAAAACAUCUGCACCUAUC ((((((((.....)))))))).....--......((((((............(((((..((..........))...)))))............))))))...... ( -14.35) >DroEre_CAF1 38809 88 - 1 AU------CAGAUGGUA-----------UCCCACUGCACACAUUAUUUUUCACUGUGCUGCUCCACCAUUUGCAUGCACAGCACCACAAAACAUCUGCACCUAUC ..------(((((((((-----------......)))...............(((((((((..........))).))))))..........))))))........ ( -19.70) >DroYak_CAF1 34018 99 - 1 GU------UACAUGGUGUAGUAAUGUUUUCCUAUGGCAAACAUUAUUUUUCACUGUGCUGCUCCACCAUUUGCAUGCACAGCGCCACAGAACAUCUGCACCUAUC ..------.....(((((((..(((((((....(((((((......)))...(((((((((..........))).)))))).)))).)))))))))))))).... ( -29.40) >DroAna_CAF1 17752 82 - 1 CU------UGGAUGGAC-----------UACCUU--CUCAAA--UUUUUUCACUGUACUUCUC--CCAUUUGCAUGCUUUGUGCGAAAAACCAUCUGCACCUAUC ..------.((((((((-----------......--......--..........)).......--...(((((((.....)))))))...))))))......... ( -12.05) >consensus AU______UAUAUAGAUUAGUAA__U__UACCAUUGCAGACAUUAUUUUUCACUGUGCUGCUCCACCAUUUGCAUGCACAGCGCCACAAAACAUCUGCACCUAUC ....................................................(((((((((..........))).))))))........................ ( -6.24 = -7.27 + 1.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:24 2006