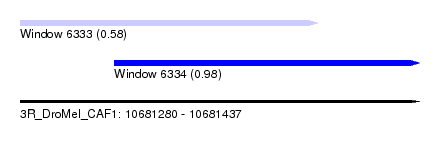
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,681,280 – 10,681,437 |
| Length | 157 |
| Max. P | 0.978070 |

| Location | 10,681,280 – 10,681,397 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10681280 117 + 27905053 UCGCCCG---CCAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGCCAAAUGCUAAGGCCAGAACGCGAAGCAUAAAGCCA ((((.((---((((.((((......))))))))))..((((((((((..((((((........))))))...((.....))..))....)))).))))......)))).((.....)).. ( -31.60) >DroSec_CAF1 22554 120 + 1 UCGCCCGCCGCCAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUGAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCA ..((((((((((((.((((......))))))))))..((((((((((..((((((........))))))..(..(.....)..)))...)))).))))......)))).))......... ( -36.30) >DroSim_CAF1 24572 120 + 1 UCGCCCGCCGCCAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCA ..((((((((((((.((((......))))))))))..((((((((((..((((((........))))))...((.....))...))...)))).))))......)))).))......... ( -37.00) >DroEre_CAF1 19863 117 + 1 UCGCCCG---ACAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCCAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCAGAGCAUAAAGCCA ..((.(.---.........(((((...)))))((((.(((((((((((.((((((........))))))...((.....))..)))...)))).))))....)))).).))......... ( -30.90) >DroYak_CAF1 14849 117 + 1 UCGCCCG---GCAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCAGAGCAUAAAGCCA ......(---((..(((..(((((...)))))((((.((((((((((..((((((........))))))...((.....))...))...)))).))))....))))....)))...))). ( -31.70) >consensus UCGCCCG___CCAGAUGAGCAGCGAUUCGCUGGCGUAGCCUAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCA .............(((((....((....((((((...))).))).....((((((........))))))..))...)))))..(((..(((((...((......))..)))))...))). (-25.42 = -25.82 + 0.40)


| Location | 10,681,317 – 10,681,437 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -20.82 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10681317 120 + 27905053 UAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGCCAAAUGCUAAGGCCAGAACGCGAAGCAUAAAGCCAAGGCUAAGCAACGGCAAAAGGCGAAAAUCCAGAAAUUCUG .....((..((((((........))))))..............(((..(((((...((......))..)))))..(((....))).......))).....)).......(((.....))) ( -21.70) >DroSec_CAF1 22594 120 + 1 UAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUGAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCAAGGCUGAGCAACGGCAAAAGGCGAAAAUCCAGAAAUUCUG ...((((..((((((........))))))..............(((..(((((...((......))..)))))...)))...)))).....((.(....).))......(((.....))) ( -24.20) >DroSim_CAF1 24612 120 + 1 UAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCAAGGCUGAGCAACGGCAAAAGGCGAAAAUCCAGAAAUUCUG (((((((..((((((........))))))...((.....))...))...)))))....(((((..((((.......(((...((((.....))))....))).....))).)...))))) ( -24.30) >DroEre_CAF1 19900 112 + 1 UAGCAGCCAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCAGAGCAUAAAGCCAAGGCUGAGCAAAGGCAA--------AAUCCAGAAAUUCUG ...(((((.((((((........))))))..............(((..(((((...((......))..)))))...)))..))))).((....))..--------....(((.....))) ( -27.90) >DroYak_CAF1 14886 112 + 1 UAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCAGAGCAUAAAGCCAAGGCUGAGCAACGGCAA--------AAUCCAGAAAUUCUG ...((((..((((((........))))))..............(((..(((((...((......))..)))))...)))...)))).((....))..--------....(((.....))) ( -24.40) >consensus UAGCAGCAAGAUUAUAAUUAAAAGUAAUCAACGACAUUAUCAAGGCAAAUGCUAAGGCCAGAACGCGGAGCAUAAAGCCAAGGCUGAGCAACGGCAAAAGGCGAAAAUCCAGAAAUUCUG ...((((..((((((........))))))..............(((..(((((...((......))..)))))...)))...)))).((....))..............(((.....))) (-20.82 = -21.22 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:18 2006