
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,678,257 – 10,678,377 |
| Length | 120 |
| Max. P | 0.999691 |

| Location | 10,678,257 – 10,678,349 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -24.36 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10678257 92 + 27905053 UUUGUCAUUGUUGCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ........((.((.(((((.(((((((((......))).((((((((.(((((......)))))..)).))))))))))))))))).)).)) ( -27.10) >DroSec_CAF1 19586 92 + 1 UUUGUCAUUGUUGCUGCGGUGAAUCAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ........((.((.(((((.((((..(((......))).((((((((.(((((......)))))..)).))))))..))))))))).)).)) ( -24.30) >DroSim_CAF1 21620 92 + 1 UUUGUCAUUGUUGCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ........((.((.(((((.(((((((((......))).((((((((.(((((......)))))..)).))))))))))))))))).)).)) ( -27.10) >DroEre_CAF1 16752 92 + 1 UUUGUCAUUGUUGCAGCGGUGAAUUAGUCAUGCCAAACCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ...............((((.((((((((........)).((((((((.(((((......)))))..)).))))))))))))))))....... ( -22.60) >DroYak_CAF1 11666 92 + 1 UUUGUCAUUGUUGCUGCGGUGAAUUAGUCAUGCCAAACCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ........((.((.(((((.((((((((........)).((((((((.(((((......)))))..)).))))))))))))))))).)).)) ( -23.80) >consensus UUUGUCAUUGUUGCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCA ........((.((.(((((.(((((((((......))).((((((((.(((((......)))))..)).))))))))))))))))).)).)) (-24.36 = -24.92 + 0.56)



| Location | 10,678,257 – 10,678,349 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -28.18 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10678257 92 - 27905053 UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGCCUGGCAUGACUAAUUCACCGCAGCAACAAUGACAAA ..((..(((((((((((((((((.(((.(((((......)))))))))))))).((...)).....)))))).)))))))............ ( -28.80) >DroSec_CAF1 19586 92 - 1 UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGCCUGGCAUGACUGAUUCACCGCAGCAACAAUGACAAA ..((..(((((((((((((((((.(((.(((((......)))))))))))))).((...)).....)))))).)))))))............ ( -28.90) >DroSim_CAF1 21620 92 - 1 UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGCCUGGCAUGACUAAUUCACCGCAGCAACAAUGACAAA ..((..(((((((((((((((((.(((.(((((......)))))))))))))).((...)).....)))))).)))))))............ ( -28.80) >DroEre_CAF1 16752 92 - 1 UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGUUUGGCAUGACUAAUUCACCGCUGCAACAAUGACAAA ..(((..(((((((((.((((((.(((.(((((......))))))))))))))((((......))))))))).)))))))............ ( -29.30) >DroYak_CAF1 11666 92 - 1 UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGUUUGGCAUGACUAAUUCACCGCAGCAACAAUGACAAA ..((..((((((((((.((((((.(((.(((((......))))))))))))))((((......))))))))).)))))))............ ( -30.00) >consensus UGGCGAUGCGGGAAUUACGAGUUUGAGGAGCAAUAACAUUUGCUUUCAACUCGGGCCUGGCAUGACUAAUUCACCGCAGCAACAAUGACAAA ..((...((((((((((((((((.(((.(((((......)))))))))))))).((...)).....)))))).)))).))............ (-28.18 = -27.78 + -0.40)

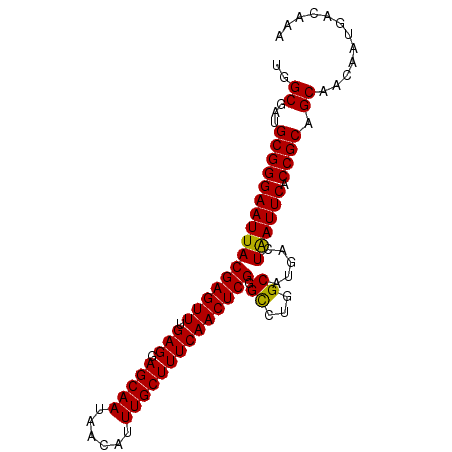

| Location | 10,678,269 – 10,678,377 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.97 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -31.78 |
| Energy contribution | -32.34 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10678269 108 + 27905053 GCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCU------------CCAGUU (((((((.(((((((((......))).((((((((.(((((......)))))..)).)))))))))))))))).((((.(((....))).))))....))).------------...... ( -34.70) >DroSec_CAF1 19598 108 + 1 GCUGCGGUGAAUCAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCU------------CCAGUU (((((((.((((..(((......))).((((((((.(((((......)))))..)).))))))..)))))))).((((.(((....))).))))....))).------------...... ( -31.90) >DroSim_CAF1 21632 108 + 1 GCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCU------------CCAGUU (((((((.(((((((((......))).((((((((.(((((......)))))..)).)))))))))))))))).((((.(((....))).))))....))).------------...... ( -34.70) >DroEre_CAF1 16764 108 + 1 GCAGCGGUGAAUUAGUCAUGCCAAACCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCU------------CCAGUU ((.((((.((((((((........)).((((((((.(((((......)))))..)).)))))))))))))))).((((.(((....))).)))).....)).------------...... ( -30.80) >DroYak_CAF1 11678 120 + 1 GCUGCGGUGAAUUAGUCAUGCCAAACCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCUCCAGCUCCAGCUCCAGUU (((((((.((((((((........)).((((((((.(((((......)))))..)).)))))))))))))))).((((.(((....))).))))...((((....)))).)))....... ( -36.00) >consensus GCUGCGGUGAAUUAGUCAUGCCAGGCCCGAGUUGAAAGCAAAUGUUAUUGCUCCUCAAACUCGUAAUUCCCGCAUCGCCAUGGUCGCGUUGCGAAUCGAGCU____________CCAGUU (((((((.(((((((((......))).((((((((.(((((......)))))..)).)))))))))))))))).((((.(((....))).))))....)))................... (-31.78 = -32.34 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:15 2006