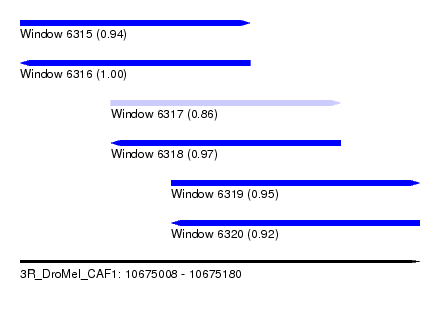
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,675,008 – 10,675,180 |
| Length | 172 |
| Max. P | 0.999913 |

| Location | 10,675,008 – 10,675,107 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -21.16 |
| Energy contribution | -20.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675008 99 + 27905053 CUGUGGCUUACUGGCC-CCAGUCAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU ...(((((..(((((.-...))))).((((.((.....)).)))).........................)))))....((((((((....)))))))). ( -26.10) >DroSec_CAF1 16393 99 + 1 CUGUGGCUUACUGGCC-UCAGUAAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU ...((((((((((...-.))))))..((((.((.....)).))))..........................))))....((((((((....)))))))). ( -25.00) >DroSim_CAF1 18411 99 + 1 CUGCGGCUUACUGGCC-UCAGUAAGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU ..(((((((((((...-.))))))..((((.((.....)).))))..........................))).))..((((((((....)))))))). ( -25.60) >DroEre_CAF1 13654 91 + 1 CUGUGGCUUACUGGGU-CCAGU-------GAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAU-UUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU ...(((((..((((((-...((-------(((.....)))))..)).))))...(((....)))...-..)))))....((((((((....)))))))). ( -30.30) >DroYak_CAF1 8455 93 + 1 CUGUGGCUUACUAGCCCCCAGU-------GAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU (((.((((....))))....((-------(((.....)))))......)))............................((((((((....)))))))). ( -24.00) >consensus CUGUGGCUUACUGGCC_CCAGU_AGAUACGAGAUACACUCACGUACACCAGAUAGAGACAAUUCAAUUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUU ..((((((.(((((...))))).......(((.....)))..............................)))).))..((((((((....)))))))). (-21.16 = -20.96 + -0.20)

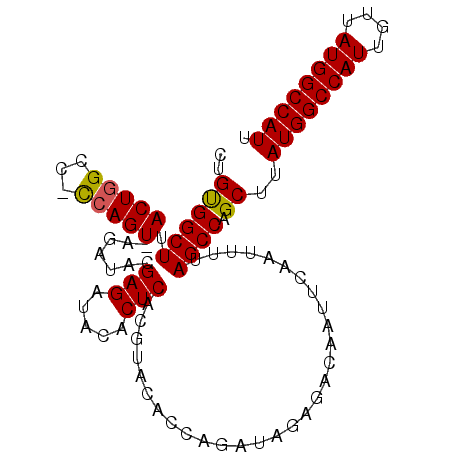
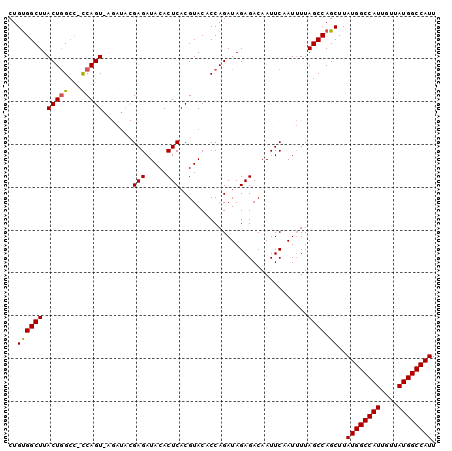
| Location | 10,675,008 – 10,675,107 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675008 99 - 27905053 AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUGACUGG-GGCCAGUAAGCCACAG .((((((((....))))))))..(((((((...........(((..(((..(((((((....)))))))..)))...)))...-)))))))......... ( -32.24) >DroSec_CAF1 16393 99 - 1 AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUUACUGA-GGCCAGUAAGCCACAG .((((((((....))))))))..(.(((((...((((....(((((.....((.(((((.((.....)).)))))))....))-))))))).)))))).. ( -31.00) >DroSim_CAF1 18411 99 - 1 AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCUUACUGA-GGCCAGUAAGCCGCAG .((((((((....))))))))..((.((((...((((....(((((.....((.(((((.((.....)).)))))))....))-))))))).)))))).. ( -32.50) >DroEre_CAF1 13654 91 - 1 AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAA-AUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUC-------ACUGG-ACCCAGUAAGCCACAG .((((((((....))))))))..(.(((((..-...((......))...((((.((..(((((.....)))-------))...-))))))..)))))).. ( -30.30) >DroYak_CAF1 8455 93 - 1 AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUC-------ACUGGGGGCUAGUAAGCCACAG .((((((((....))))))))..(.(((((...((((....(((((((...(((((((....)))))))..-------..))))))))))).)))))).. ( -31.20) >consensus AAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAAUUGAAUUGUCUCUAUCUGGUGUACGUGAGUGUAUCUCGUAUCU_ACUGG_GGCCAGUAAGCCACAG .((((((((....))))))))..(.(((((.....................(((((((....))))))).........((((.....)))).)))))).. (-28.66 = -28.34 + -0.32)

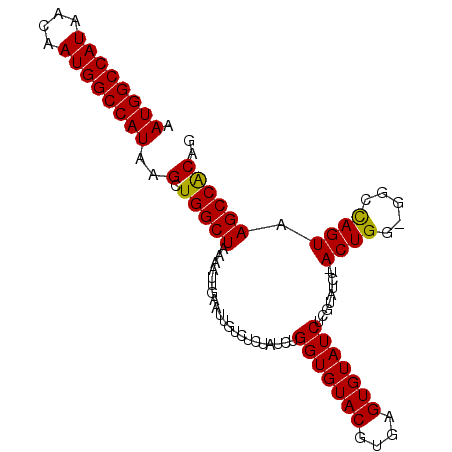
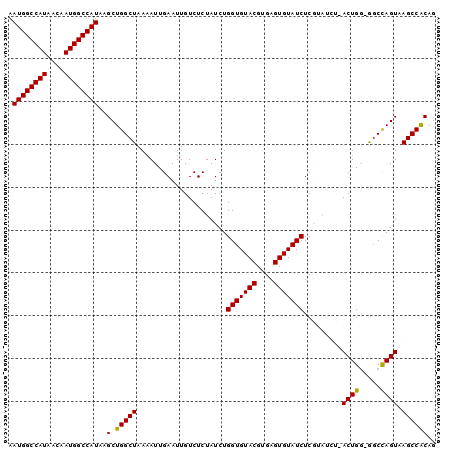
| Location | 10,675,047 – 10,675,146 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675047 99 + 27905053 A-----CGUACACCAG--AUAGAGACAAUUCAA-------------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUG .-----..........--...............-------------.....((((((..((((((((....))))))))......((((.......-......))))...)))))).... ( -23.62) >DroSec_CAF1 16432 99 + 1 A-----CGUACACCAG--AUAGAGACAAUUCAA-------------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUG .-----..........--...............-------------.....((((((..((((((((....))))))))......((((.......-......))))...)))))).... ( -23.62) >DroSim_CAF1 18450 99 + 1 A-----CGUACACCAG--AUAGAGACAAUUCAA-------------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUG .-----..........--...............-------------.....((((((..((((((((....))))))))......((((.......-......))))...)))))).... ( -23.62) >DroEre_CAF1 13686 98 + 1 A-----CGUACACCAG--AUAGAGACAAUUCAA-------------U-UUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUG .-----..........--...............-------------.-...((((((..((((((((....))))))))......((((.......-......))))...)))))).... ( -23.62) >DroWil_CAF1 20973 113 + 1 AACAUUCGGUCAGCUGCUAUAUAAACAAUUCAAUAAUAUACAAAAAUUUUAGACAGUU-------GUUGUUGCUGCUGCUGUUGCUGACUUACGUUUUCAGCCAAUAGCCACAAACGACA ........((((((.((......(((((((...(((.((......)).)))...))))-------)))...)).)))(((((((((((.........)))).))))))).......))). ( -24.30) >DroYak_CAF1 8488 99 + 1 A-----CGUACACCAG--AUAGAGACAAUUCAA-------------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUGGCUAACAUU-UGAACCGCUGACCGUUGGCGUUG .-----...((.((((--....((....(((((-------------..((((((((...((((((((....)))))))).....))))))))...)-))))...)).....)))).)).. ( -31.80) >consensus A_____CGUACACCAG__AUAGAGACAAUUCAA_____________UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU_UCAACCGCUGACCGUUGGCGUUG ...................................................((((((..((((((((....))))))))......((((..............))))...)))))).... (-17.28 = -17.92 + 0.64)

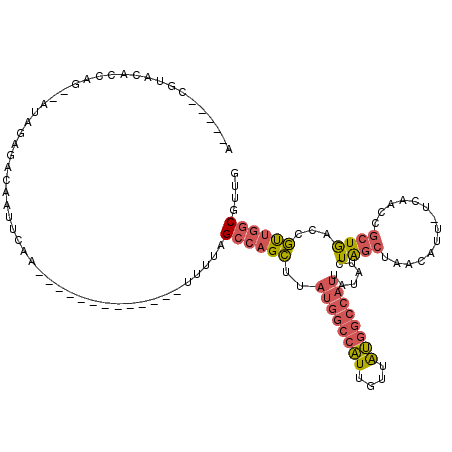
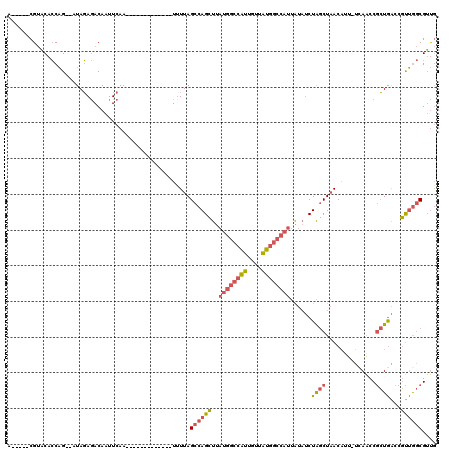
| Location | 10,675,047 – 10,675,146 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675047 99 - 27905053 CAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA-------------UUGAAUUGUCUCUAU--CUGGUGUACG-----U ..((((((..((...((((((.(-(...((((((((...(.((((((((....)))))))).).))))))))..-------------)).))))))..))..--.))))))...-----. ( -34.80) >DroSec_CAF1 16432 99 - 1 CAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA-------------UUGAAUUGUCUCUAU--CUGGUGUACG-----U ..((((((..((...((((((.(-(...((((((((...(.((((((((....)))))))).).))))))))..-------------)).))))))..))..--.))))))...-----. ( -34.80) >DroSim_CAF1 18450 99 - 1 CAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA-------------UUGAAUUGUCUCUAU--CUGGUGUACG-----U ..((((((..((...((((((.(-(...((((((((...(.((((((((....)))))))).).))))))))..-------------)).))))))..))..--.))))))...-----. ( -34.80) >DroEre_CAF1 13686 98 - 1 CAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAA-A-------------UUGAAUUGUCUCUAU--CUGGUGUACG-----U ..((((((..((...((((((.(-(...((((((((...(.((((((((....)))))))).).))))))))-.-------------)).))))))..))..--.))))))...-----. ( -34.40) >DroWil_CAF1 20973 113 - 1 UGUCGUUUGUGGCUAUUGGCUGAAAACGUAAGUCAGCAACAGCAGCAGCAACAAC-------AACUGUCUAAAAUUUUUGUAUAUUAUUGAAUUGUUUAUAUAGCAGCUGACCGAAUGUU ...(((((.((((.....)))).)))))...((((((....((....))......-------..((((.((((...((((........))))...)))).))))..))))))........ ( -21.10) >DroYak_CAF1 8488 99 - 1 CAACGCCAACGGUCAGCGGUUCA-AAUGUUAGCCAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA-------------UUGAAUUGUCUCUAU--CUGGUGUACG-----U ..((((((..((...((((((((-(...((((((((...(.((((((((....)))))))).).))))))))..-------------)))))))))..))..--.))))))...-----. ( -41.20) >consensus CAACGCCAACGGUCAGCGGUUGA_AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA_____________UUGAAUUGUCUCUAU__CUGGUGUACG_____U ..((((((..(((((((((((((.....)))))).......((((((((....))))))))..)))))))...................((....))........))))))......... (-25.27 = -25.25 + -0.02)

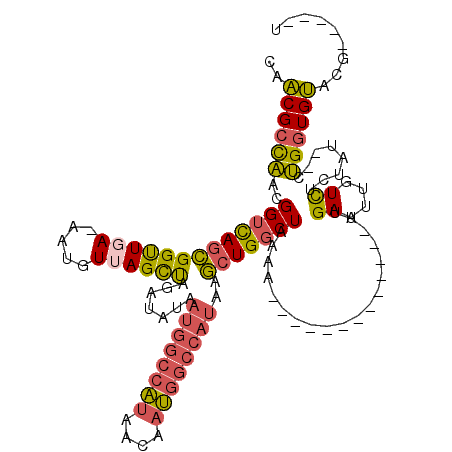

| Location | 10,675,073 – 10,675,180 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -18.86 |
| Energy contribution | -20.42 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675073 107 + 27905053 ------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUGGC------GAAACAAAAUGAAAUAAAACGCUCCACACUCG ------.....((((((..((((((((....))))))))........((((((...-(((.....)))..))))))))))))------................................ ( -30.00) >DroSec_CAF1 16458 107 + 1 ------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUGGC------GAAACAAAAUGAAAUAAAACGCUACACACUCG ------.....((((((..((((((((....))))))))........((((((...-(((.....)))..))))))))))))------................................ ( -30.00) >DroSim_CAF1 18476 107 + 1 ------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUGGC------GAAACAAAAUGAAAUAAAACGCUCCACACUCG ------.....((((((..((((((((....))))))))........((((((...-(((.....)))..))))))))))))------................................ ( -30.00) >DroEre_CAF1 13712 106 + 1 ------U-UUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU-UCAACCGCUGACCGUUGGCGUUGGC------GAAACAAAAUGAAAUAAAACGCUUCACACUCG ------.-...((((((..((((((((....))))))))........((((((...-(((.....)))..))))))))))))------................................ ( -30.00) >DroWil_CAF1 21013 113 + 1 CAAAAAUUUUAGACAGUU-------GUUGUUGCUGCUGCUGUUGCUGACUUACGUUUUCAGCCAAUAGCCACAAACGACAACAACAACGAAUAACAAAAAAAUAAAGCCCACCACAUUUG ...............(((-------(((((((.((..(((((((((((.........)))).)))))))..))..))))))))))................................... ( -25.90) >DroYak_CAF1 8514 107 + 1 ------UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUGGCUAACAUU-UGAACCGCUGACCGUUGGCGUUGGC------AAAACAAAAUGAAAUAAAACGCUCCACACUCG ------..((((((((...((((((((....)))))))).....))))))))((((-(.....((..(((....).))..))------......)))))..................... ( -32.20) >consensus ______UUUUAGCCAGCUUAUGGCCAUUGUUAUGGCCAUUAUAUCUAGCUAACAUU_UCAACCGCUGACCGUUGGCGUUGGC______GAAACAAAAUGAAAUAAAACGCUCCACACUCG ...........((((((..((((((((....))))))))........((((((....(((.....)))..))))))))))))...................................... (-18.86 = -20.42 + 1.56)
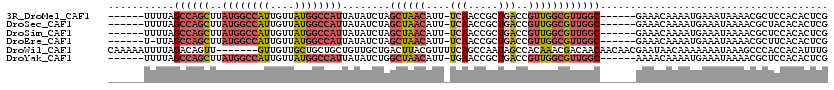
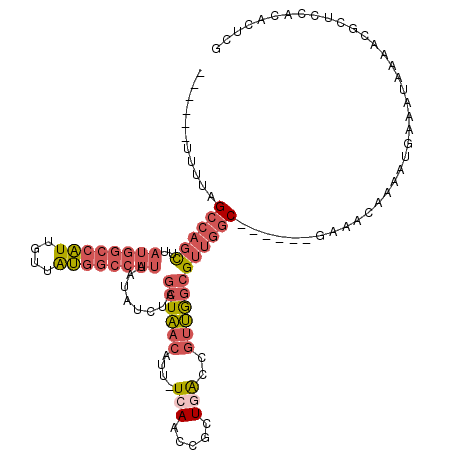

| Location | 10,675,073 – 10,675,180 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10675073 107 - 27905053 CGAGUGUGGAGCGUUUUAUUUCAUUUUGUUUC------GCCAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA------ ...(.((((((((.............))))))------))).....((((.(....).)))).-....((((((((...(.((((((((....)))))))).).))))))))..------ ( -33.32) >DroSec_CAF1 16458 107 - 1 CGAGUGUGUAGCGUUUUAUUUCAUUUUGUUUC------GCCAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA------ .(.((((...(((...((........))...)------))..)))))...(((((((((((((-....)))))).......((((((((....))))))))..)))))))....------ ( -32.70) >DroSim_CAF1 18476 107 - 1 CGAGUGUGGAGCGUUUUAUUUCAUUUUGUUUC------GCCAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA------ ...(.((((((((.............))))))------))).....((((.(....).)))).-....((((((((...(.((((((((....)))))))).).))))))))..------ ( -33.32) >DroEre_CAF1 13712 106 - 1 CGAGUGUGAAGCGUUUUAUUUCAUUUUGUUUC------GCCAACGCCAACGGUCAGCGGUUGA-AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAA-A------ ...(.((((((((.............))))))------))).....((((.(....).)))).-....((((((((...(.((((((((....)))))))).).))))))))-.------ ( -33.82) >DroWil_CAF1 21013 113 - 1 CAAAUGUGGUGGGCUUUAUUUUUUUGUUAUUCGUUGUUGUUGUCGUUUGUGGCUAUUGGCUGAAAACGUAAGUCAGCAACAGCAGCAGCAACAAC-------AACUGUCUAAAAUUUUUG ((((.((..(((((...........(.....)(((((((((((....(((.(((....(((((...(....))))))...))).)))))))))))-------))).)))))..)).)))) ( -31.00) >DroYak_CAF1 8514 107 - 1 CGAGUGUGGAGCGUUUUAUUUCAUUUUGUUUU------GCCAACGCCAACGGUCAGCGGUUCA-AAUGUUAGCCAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA------ .((.(((.(.(((((......(.....)....------...)))))).))).)).........-....((((((((...(.((((((((....)))))))).).))))))))..------ ( -32.72) >consensus CGAGUGUGGAGCGUUUUAUUUCAUUUUGUUUC______GCCAACGCCAACGGUCAGCGGUUGA_AAUGUUAGCUAGAUAUAAUGGCCAUAACAAUGGCCAUAAGCUGGCUAAAA______ .((.(((...(((((..........................)))))..))).))..............((((((((.....((((((((....))))))))...))))))))........ (-21.80 = -22.47 + 0.67)

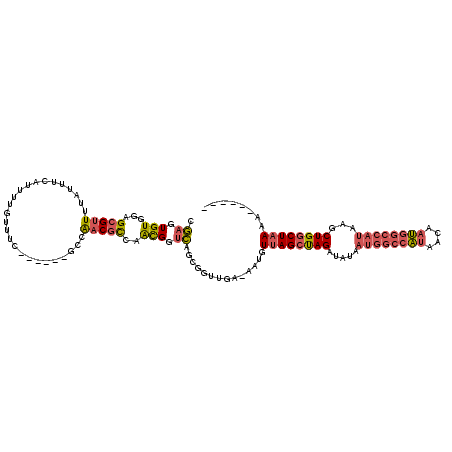
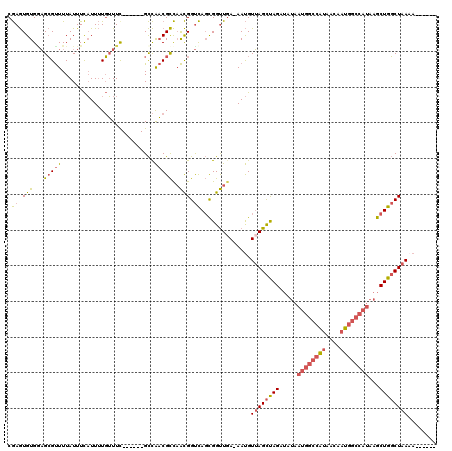
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:04 2006