
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,667,979 – 10,668,085 |
| Length | 106 |
| Max. P | 0.970474 |

| Location | 10,667,979 – 10,668,085 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -11.76 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
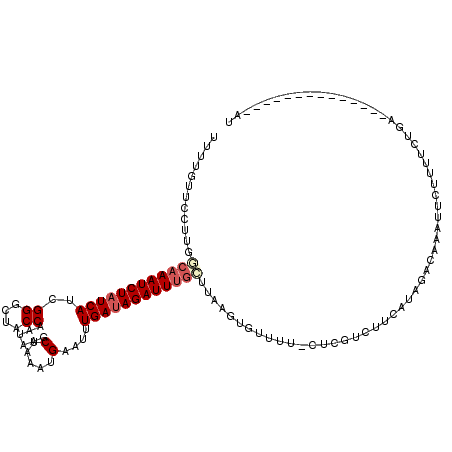
>3R_DroMel_CAF1 10667979 106 + 27905053 UUUUGUUCCUUAACAAAUCUAUCAUCGGGCUACCAAUAAGCUAAAUGUAUUUGAUAGAUUUGUUUAAGUGUUUUCCUAGUCUUCGUAGACAAAUUAUUUUCUGA--------------AU ....((((((((((((((((((((..((....)).....((.....))...))))))))))).)))))..........((((....))))............))--------------)) ( -22.50) >DroSec_CAF1 9581 106 + 1 UUUCGUUCCCUGACAAAUCUAUCAUAGGGCUACCAAUAAGCUCAAUGAAUUUGAUAGAUUUGCUUAAGUGUUUUACUCGUCUUCAUAGACAAAUUCUUUUCUGA--------------AU ..........((((((((((((((..(((((.......)))))........))))))))))).)))............((((....))))..((((......))--------------)) ( -20.40) >DroSim_CAF1 9712 106 + 1 UUUCGUUCCUUGGCAAAUCUAUCAUCGGGCUACCAAUAAGCUAAAUGAAUUUGAUAGAUUUGCUUAAGUGUUUUCCUCGUCUUCAUAGACAAAUUCUUUUCUGA--------------AU ........(((((((((((((((((((((((.......))))...)))...)))))))))))).))))..........((((....))))..((((......))--------------)) ( -25.30) >DroEre_CAF1 6510 117 + 1 UUCUGUUCCUUUGCAAAUCUAUCAUCGGGCUACCAAAAAACUAAUUGAAUCUGAUAGAUUUGC-AAAGUGUUU--CUUUUUUGCUUCAACAAAUUCCUUGCUGAGUCCCAGUACAUGAGU ........((((((((((((((((..((....))......(.....)....))))))))))))-)))).....--.................((((..(((((.....)))))...)))) ( -27.20) >DroYak_CAF1 1150 105 + 1 UUCUGUUCCUUGGAAAAUCUUUCAUCGGGCUACCAAGAAACUAAAUGAAUCUGAUAGAUUCG------UUUUU--CUUUUCUA-------AAGCUACUUUCUGAAUCCCAGUACAUGAGU ((((((..((.(((...((........((((....(((((..(((((((((.....))))))------)))..--..))))).-------.)))).......)).))).)).))).))). ( -19.16) >consensus UUUUGUUCCUUGGCAAAUCUAUCAUCGGGCUACCAAUAAGCUAAAUGAAUUUGAUAGAUUUGCUUAAGUGUUUU_CUCGUCUUCAUAGACAAAUUCUUUUCUGA______________AU ............((((((((((((..((....))......(.....)....))))))))))))......................................................... (-11.76 = -12.60 + 0.84)


| Location | 10,667,979 – 10,668,085 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.59 |
| Mean single sequence MFE | -23.71 |
| Consensus MFE | -13.50 |
| Energy contribution | -14.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
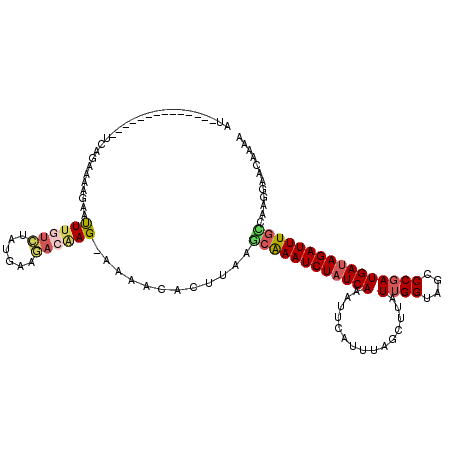
>3R_DroMel_CAF1 10667979 106 - 27905053 AU--------------UCAGAAAAUAAUUUGUCUACGAAGACUAGGAAAACACUUAAACAAAUCUAUCAAAUACAUUUAGCUUAUUGGUAGCCCGAUGAUAGAUUUGUUAAGGAACAAAA .(--------------((............((((....))))..........(((((.(((((((((((...............((((....)))))))))))))))))))))))..... ( -20.36) >DroSec_CAF1 9581 106 - 1 AU--------------UCAGAAAAGAAUUUGUCUAUGAAGACGAGUAAAACACUUAAGCAAAUCUAUCAAAUUCAUUGAGCUUAUUGGUAGCCCUAUGAUAGAUUUGUCAGGGAACGAAA .(--------------((........((((((((....))))))))......(((..((((((((((((.....((..(.....)..)).......))))))))))))..)))...))). ( -23.60) >DroSim_CAF1 9712 106 - 1 AU--------------UCAGAAAAGAAUUUGUCUAUGAAGACGAGGAAAACACUUAAGCAAAUCUAUCAAAUUCAUUUAGCUUAUUGGUAGCCCGAUGAUAGAUUUGCCAAGGAACGAAA .(--------------((.........(((((((....))))))).......(((..((((((((((((...............((((....))))))))))))))))..)))...))). ( -24.06) >DroEre_CAF1 6510 117 - 1 ACUCAUGUACUGGGACUCAGCAAGGAAUUUGUUGAAGCAAAAAAG--AAACACUUU-GCAAAUCUAUCAGAUUCAAUUAGUUUUUUGGUAGCCCGAUGAUAGAUUUGCAAAGGAACAGAA .........(((.(..((((((((...))))))))..).......--.....((((-((((((((((((......(((((....))))).......))))))))))))))))...))).. ( -32.12) >DroYak_CAF1 1150 105 - 1 ACUCAUGUACUGGGAUUCAGAAAGUAGCUU-------UAGAAAAG--AAAAA------CGAAUCUAUCAGAUUCAUUUAGUUUCUUGGUAGCCCGAUGAAAGAUUUUCCAAGGAACAGAA ..((.(((.((((((((((....((.(((.-------.(((((..--(((..------.(((((.....))))).)))..))))).))).))....)))).....)))).))..))))). ( -18.40) >consensus AU______________UCAGAAAAGAAUUUGUCUAUGAAGACAAG_AAAACACUUAAGCAAAUCUAUCAAAUUCAUUUAGCUUAUUGGUAGCCCGAUGAUAGAUUUGCCAAGGAACAAAA ...........................((((((......))))))............((((((((((((...............((((....))))))))))))))))............ (-13.50 = -14.30 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:43 2006