
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,664,236 – 10,664,472 |
| Length | 236 |
| Max. P | 0.998467 |

| Location | 10,664,236 – 10,664,340 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -22.08 |
| Energy contribution | -22.64 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10664236 104 - 27905053 CUCUUUUAAUGUUUUCAUAACUUUCAGCGCGUCUGUCAAUAAGUCGAAUUCUUGGAAACAA-GCUCGAAACUUAAGUCACGUUACGUGACUGCCAUGUCAAUGGC--------------- .................(((.(((((((.((.((.......)).)).....(((....)))-))).)))).)))(((((((...)))))))(((((....)))))--------------- ( -25.50) >DroSec_CAF1 5892 102 - 1 C--UUUUAAUGUUUUCAUAACUUUCGGCGCGUCUGUCAAUAAGUCGAAUUCUUGGAAACAA-GCUCGAAACUUAAGUCACGUUACGUGACUGCCAUGUCAAUGGC--------------- .--......((((((((.....((((((..............))))))....)))))))).-............(((((((...)))))))(((((....)))))--------------- ( -27.64) >DroSim_CAF1 5952 102 - 1 C--UUUUAAUGUUUUCAUAACUUUCGACGCGUCUGUCAAUAAGUCGAAUUCUUGGAAACAA-GCUCGAAACUUAAGUCACGUUACGUGACUGCCAUGUCAAUGGC--------------- .--......((((((((.....((((((..............))))))....)))))))).-............(((((((...)))))))(((((....)))))--------------- ( -28.24) >DroEre_CAF1 2379 102 - 1 C--UUUUAAUGUUUUCAUAACUUUCGGCGCGUCUGUCAAUAAGUCGAAUUCUUGGAAACAG-GCUCGAAACUUAAGUCACGUUACGUGACUCCCAUGCCAAUGGC--------------- .--......((((((((.....((((((..............))))))....))))))))(-((..........(((((((...))))))).....)))......--------------- ( -24.60) >DroWil_CAF1 11858 116 - 1 ----GCUAAUGUUUUCAUAACUUUUGGCGCGUCUGUCAAUAAGUUGAAUUCUUGGAAACAAAAUUCUAAACUUAAGUCAAGUUACGUGACUUUCAUGUCAAUGCCCUAGGCAGGGACCUG ----(((((.(((.....)))..)))))...........(((((((((((.(((....))))))))..))))))(((((.......)))))..((.(((..((((...))))..))).)) ( -27.40) >consensus C__UUUUAAUGUUUUCAUAACUUUCGGCGCGUCUGUCAAUAAGUCGAAUUCUUGGAAACAA_GCUCGAAACUUAAGUCACGUUACGUGACUGCCAUGUCAAUGGC_______________ .........((((((((.....((((((..............))))))....))))))))..............(((((((...)))))))(((((....)))))............... (-22.08 = -22.64 + 0.56)



| Location | 10,664,340 – 10,664,447 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -22.65 |
| Energy contribution | -23.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10664340 107 + 27905053 GAACGCCCACCCCCCAAAUGCCUGGCAAGCUUUGGGGUUUUUUCUCCUUUU--GGUAUGGCCAAAA-----AAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGC ((((.......(((((((.((.......)))))))))((((((....((((--((.....))))))-----..)))))).)))).....(((((((..(....)..))))))). ( -32.30) >DroSec_CAF1 5994 112 + 1 GAACGGCCUCCCCCCAAAUGCCUGGCAAGCUUUGGGGUUUUUUCUCCUUUU--GGUAUGGCCAAAAAAAGAAAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUACCCAGC ((((.......(((((((.((.......)))))))))((((((((..((((--((.....))))))..))))))))....)))).....(((((..............))))). ( -33.54) >DroSim_CAF1 6054 111 + 1 GAACGCCCUCCCCCCAAAUGCCUGGCAAGCUUUGGGGUUUUUUCUCCUUUU--GGUAUGGCCAAA-AAAGAAAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGC ((((.......(((((((.((.......)))))))))((((((((..((((--((.....)))))-).))))))))....)))).....(((((((..(....)..))))))). ( -38.00) >DroEre_CAF1 2481 103 + 1 GAACGCCCUCUCCCCAAAUGCCUGGCAAGCUUUGG-GUUUUUUCUCCAUACCAAAUAUGACCA----------AAAAAAGGUUCACAGCCUGGGCAAAAAUGAUGGUGCCCAGU ((((........((((((.((.......)))))))-)((((((...((((.....))))...)----------)))))..)))).....(((((((..(....)..))))))). ( -25.20) >consensus GAACGCCCUCCCCCCAAAUGCCUGGCAAGCUUUGGGGUUUUUUCUCCUUUU__GGUAUGGCCAAA______AAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGC ((((.......(((((((.((.......)))))))))................((.....))..................)))).....(((((((..(....)..))))))). (-22.65 = -23.65 + 1.00)


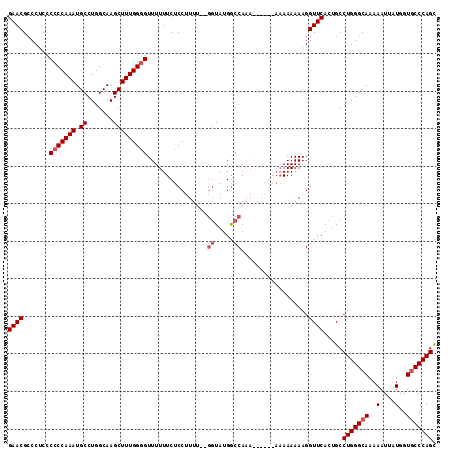
| Location | 10,664,340 – 10,664,447 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -39.45 |
| Consensus MFE | -27.34 |
| Energy contribution | -30.53 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.939021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10664340 107 - 27905053 GCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUU-----UUUUGGCCAUACC--AAAAGGAGAAAAAACCCCAAAGCUUGCCAGGCAUUUGGGGGGUGGGCGUUC .(((((((..........)))))))...((..(((((((((((-----((((((.....))--))))))))))))..((((((((((.....))).))))))))))..)).... ( -42.30) >DroSec_CAF1 5994 112 - 1 GCUGGGUACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUUUCUUUUUUUGGCCAUACC--AAAAGGAGAAAAAACCCCAAAGCUUGCCAGGCAUUUGGGGGGAGGCCGUUC .(((((((..........)))))))........(((((((((((((((((((((.....))--))))))))))))))((((((((((.....))).))))))))))))...... ( -42.60) >DroSim_CAF1 6054 111 - 1 GCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUUUCUUU-UUUGGCCAUACC--AAAAGGAGAAAAAACCCCAAAGCUUGCCAGGCAUUUGGGGGGAGGGCGUUC .(((((((..........)))))))........(((((((((((((((-(((((.....))--)))).)))))))))((((((((((.....))).))))))))))))...... ( -41.20) >DroEre_CAF1 2481 103 - 1 ACUGGGCACCAUCAUUUUUGCCCAGGCUGUGAACCUUUUUU----------UGGUCAUAUUUGGUAUGGAGAAAAAAC-CCAAAGCUUGCCAGGCAUUUGGGGAGAGGGCGUUC .(((((((..........)))))))(((.(.....((((((----------(..((((((...)))))).)))))))(-((((((((.....))).))))))...).))).... ( -31.70) >consensus GCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUU______UUUGGCCAUACC__AAAAGGAGAAAAAACCCCAAAGCUUGCCAGGCAUUUGGGGGGAGGGCGUUC .(((((((..........))))))).....((((.((((((((((((((...((.....))..))))))))))))))((((((((((.....))).))))))).......)))) (-27.34 = -30.53 + 3.19)



| Location | 10,664,380 – 10,664,472 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -25.31 |
| Consensus MFE | -18.38 |
| Energy contribution | -19.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10664380 92 + 27905053 UUUCUCCUUUU--GGUAUGGCCAAAA-----AAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGCAAAAGGGAGAGAAGAA-------GAAAUGUGA (((((((((((--(((((((((....-----........)).))).))))((((((..(....)..))))))...)))))))))))....-------......... ( -27.30) >DroSec_CAF1 6034 97 + 1 UUUCUCCUUUU--GGUAUGGCCAAAAAAAGAAAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUACCCAGCAAAAGGGAAAGAAGGA-------GAAAUGUGA (((((((((((--......(((................((((.....)))).))).............(((.......)))..)))))))-------))))..... ( -23.63) >DroSim_CAF1 6094 96 + 1 UUUCUCCUUUU--GGUAUGGCCAAA-AAAGAAAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGCAAAAGGGAAAGAUGGA-------GAAAUGUGA (((((((((((--((.....)))))-)..............(((.((..(((((((..(....)..)))))))....)).)))....)))-------))))..... ( -27.20) >DroEre_CAF1 2520 96 + 1 UUUCUCCAUACCAAAUAUGACCA----------AAAAAAGGUUCACAGCCUGGGCAAAAAUGAUGGUGCCCAGUAAAAGGGAGAGAAGGCGGAAUGUGAAAUGUGA (((((((...........((((.----------......))))......(((((((..(....)..)))))))......))))))).................... ( -23.10) >consensus UUUCUCCUUUU__GGUAUGGCCAAA______AAAAAAAAGGUUCACUGCCUGGGCAAAAAUUAUGGUGCCCAGCAAAAGGGAAAGAAGGA_______GAAAUGUGA (((((((((((...((......................((((.....))))(((((..(....)..))))).)).))))))))))).................... (-18.38 = -19.50 + 1.12)



| Location | 10,664,380 – 10,664,472 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -16.64 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10664380 92 - 27905053 UCACAUUUC-------UUCUUCUCUCCCUUUUGCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUU-----UUUUGGCCAUACC--AAAAGGAGAAA .........-------......(((((...((((((((((..........)))))).))))..............-----((((((.....))--))))))))).. ( -23.70) >DroSec_CAF1 6034 97 - 1 UCACAUUUC-------UCCUUCUUUCCCUUUUGCUGGGUACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUUUCUUUUUUUGGCCAUACC--AAAAGGAGAAA .....((((-------(((...........((((((((((..........)))))).))))...................((((((.....))--))))))))))) ( -23.30) >DroSim_CAF1 6094 96 - 1 UCACAUUUC-------UCCAUCUUUCCCUUUUGCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUUUCUUU-UUUGGCCAUACC--AAAAGGAGAAA .....((((-------(((...........((((((((((..........)))))).))))..................(-(((((.....))--))))))))))) ( -25.80) >DroEre_CAF1 2520 96 - 1 UCACAUUUCACAUUCCGCCUUCUCUCCCUUUUACUGGGCACCAUCAUUUUUGCCCAGGCUGUGAACCUUUUUU----------UGGUCAUAUUUGGUAUGGAGAAA .....((((.(((.((.................(((((((..........)))))))..(((((.((......----------.)))))))...)).))))))).. ( -23.60) >consensus UCACAUUUC_______UCCUUCUCUCCCUUUUGCUGGGCACCAUAAUUUUUGCCCAGGCAGUGAACCUUUUUUUU______UUUGGCCAUACC__AAAAGGAGAAA ......................(((((......(((((((..........)))))))...(((..((.................)).))).........))))).. (-16.64 = -16.20 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:33 2006