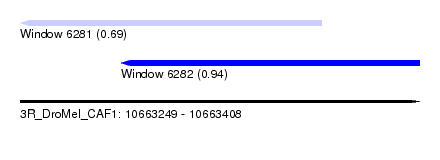
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,663,249 – 10,663,408 |
| Length | 159 |
| Max. P | 0.940930 |

| Location | 10,663,249 – 10,663,369 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.62 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -15.36 |
| Energy contribution | -16.55 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10663249 120 - 27905053 AUUCCAAAAUGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUUUGUUUAAGAAUAUUUGUAUCCUUUCUAGAUUGCCUGUAGACAUUUAAGGUU ..(((.....)))(((((((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))))))))........((((((((.(.....).)))).....)))).. ( -24.20) >DroSec_CAF1 4905 114 - 1 AUUCCUGAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUGAGAAU------AUCCUUUUUAGAUUGCCUGUAGACAUUUAAGGUU ....(((((.((((((((((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))))))..------))))).)))))...((((...........)))). ( -32.50) >DroSim_CAF1 4952 120 - 1 AUUCCUAAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUAUGAAUAUUUGUAUCCUUUUUAGAUGGCCUGUAGACAUUUAAGGUU ....(((((.(((((((.((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))).))........))))).)))))..(((((...........))))) ( -26.70) >DroEre_CAF1 1398 113 - 1 GUAUUAAA--AAAAUUCUUAAAUUUCUUAAAGGGAAACGAAGCUUAUACCAAGUUAAUAACUAGAUGGGUAAGGGAAAACAU---UAUCUUUUUUAGAUAGCCU--UGGCUAUUAAGGUU ........--.....((((((...(((.(((((((.......((..((((...(((.....)))...))))..)).......---..))))))).)))((((..--..)))))))))).. ( -18.49) >consensus AUUCCAAAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUAAGAAUAU___UAUCCUUUUUAGAUUGCCUGUAGACAUUUAAGGUU ....(((((.((((((((((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))))))........))))).)))))...((((...........)))). (-15.36 = -16.55 + 1.19)



| Location | 10,663,289 – 10,663,408 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.10 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10663289 119 - 27905053 A-AAGUAUCGUAUUUUCAAAAGCCUAUCCCUUCUUUGAACAUUCCAAAAUGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUUUGUUUAAGAAU .-........................((((...((((.......))))..))))((((((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))))))). ( -25.10) >DroSec_CAF1 4939 119 - 1 A-AAGUAUCAUAUUUUCAAAGGCCUAUCCCUUCUUUGAACAUUCCUGAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUGAGAAU .-............((((((((.........))))))))...((((....))))((((((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))))))). ( -29.70) >DroSim_CAF1 4992 119 - 1 A-AAGUAUCAUAUUUUCAAAGGCCGAUCCCUUCUUUGAACAUUCCUAAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUAUGAAU .-............((((((((.........))))))))...((((....))))(((.((((((.(((((..(..(((.(((((......))))).)))..)..))))))))))).))). ( -24.60) >DroEre_CAF1 1433 118 - 1 AAAAGUAUCGCAUUUUCAAAGGGCUAUCCGUUCUUUGAAGGUAUUAAA--AAAAUUCUUAAAUUUCUUAAAGGGAAACGAAGCUUAUACCAAGUUAAUAACUAGAUGGGUAAGGGAAAAC ..((((.(((.(((((((((((((.....)))))))))))))......--............(((((.....)))))))).)))).((((...(((.....)))...))))......... ( -23.30) >consensus A_AAGUAUCAUAUUUUCAAAGGCCUAUCCCUUCUUUGAACAUUCCAAAACGGGAUUCUUAAACAUGAUAUUACAGCAUAAAGUUUAUAACAGCUUAAUGAAGCGAUGUCUGUUUAAGAAU ..............((((((((.........)))))))).(((((.....)))))(((((((((.(((((..(..(((.(((((......))))).)))..)..)))))))))))))).. (-15.96 = -16.40 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:27 2006