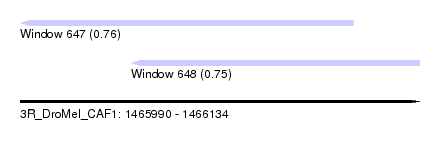
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,465,990 – 1,466,134 |
| Length | 144 |
| Max. P | 0.762732 |

| Location | 1,465,990 – 1,466,110 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -26.75 |
| Energy contribution | -27.75 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
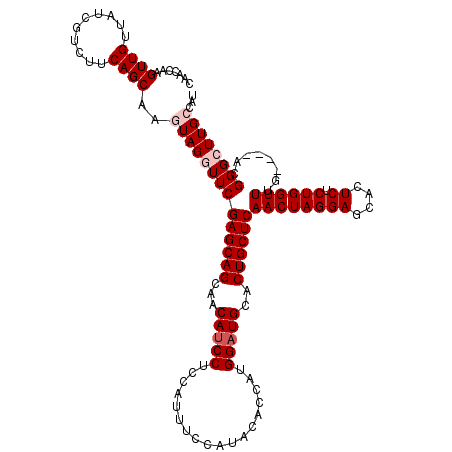
>3R_DroMel_CAF1 1465990 120 - 27905053 CAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUCAACUAGGAGCACUCUCUGGUUUGACUGACGGGCUUGCCAU .......((((..........))))..((((((((((((((...(((((..................)))))..))))))((((((((....)).))))))........))))))))... ( -32.47) >DroSec_CAF1 38536 116 - 1 CAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAGCAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUCAACUAGGAGCACUCUCUGGUUUG----ACGGGCUUGCCAU .......((((..........))))..((((((((((((((..((((((..................)))))).))))))((((((((....)).))))))..----..))))))))... ( -36.07) >DroSim_CAF1 36813 116 - 1 CAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAGCAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUCAACUAGGAGCACUCUCUGGUUUG----ACGGGCUUGCCAU .......((((..........))))..((((((((((((((..((((((..................)))))).))))))((((((((....)).))))))..----..))))))))... ( -36.07) >DroEre_CAF1 40258 116 - 1 CAACCAAGUUGUUAUCGUCUUCAGCAAAUAGAUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUCAACUAGGAGCAUUCUCUGGUUUG----ACGGGCUUGCCAU ....(((((.....(((((................((((((...(((((..................)))))..))))))(((((((......).)))))).)----))))))))).... ( -28.37) >DroYak_CAF1 38659 116 - 1 CAACCAAGUUGUUAUCGUCUUCAGCAAAUAGAUUCGAGCAGCAACACCCUCCAUUUCCAUACACUAUGGAUGCACUGCUCAACUAGGAGCAUUCUCUGGUUUG----AAGGGCUUGCCAU .......((((((.(((...((........))..)))..)))))).((((.((..((((((...)))))).....(((((......)))))..........))----.))))........ ( -26.40) >consensus CAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUCAACUAGGAGCACUCUCUGGUUUG____ACGGGCUUGCCAU .......((((..........))))..((((((((((((((...(((((..................)))))..))))))((((((((....)).))))))........))))))))... (-26.75 = -27.75 + 1.00)

| Location | 1,466,030 – 1,466,134 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.04 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
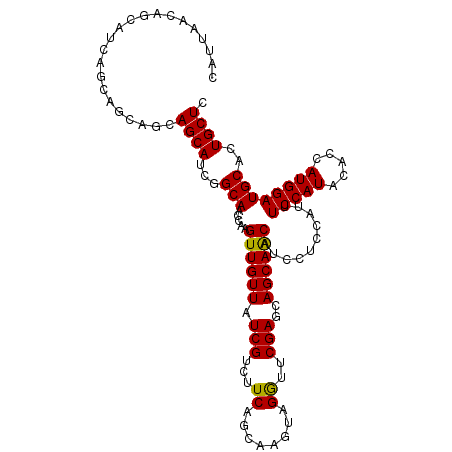
>3R_DroMel_CAF1 1466030 104 - 27905053 CAUUAACAGCAU------CAGCAGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUC ........((..------..)).((....))((((.(.(((((.........))))).).)))).((((((...(((((..................)))))..)))))) ( -21.77) >DroSec_CAF1 38572 110 - 1 CAUUAACAGCAUCAGCAUCAGCAGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAGCAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUC ...(((((((....((....)).((....))......))))))).....................((((((..((((((..................)))))).)))))) ( -26.97) >DroSim_CAF1 36849 110 - 1 CAUUAACAGCAUCAGCAUCAGCAGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAGCAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUC ...(((((((....((....)).((....))......))))))).....................((((((..((((((..................)))))).)))))) ( -26.97) >DroEre_CAF1 40294 110 - 1 CACUAACAGCAACAUCAGCAUCAGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAAUAGAUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUC ...(((((((.......((....))...(....)...)))))))..((((.........))))..((((((...(((((..................)))))..)))))) ( -22.77) >DroYak_CAF1 38695 104 - 1 AAUUGACA---ACAUCAGC---AGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAAUAGAUUCGAGCAGCAACACCCUCCAUUUCCAUACACUAUGGAUGCACUGCUC ........---.....(((---((.....(((.....((((((.(((...((........))..)))..))))))..........((((((...))))))))).))))). ( -22.10) >consensus CAUUAACAGCAUCAGCAGCAGCAGCAUCGGCAACCAAGUUGUUAUCGUCUUCAGCAAGUAGGUUCGAGCAGCAACAUCCUCCAUUUCCAUACACCAUGGAUGCACUGCUC ......................((((...(((.....((((((.(((...((........))..)))..))))))..........(((((.....))))))))..)))). (-20.52 = -20.04 + -0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:08 2006