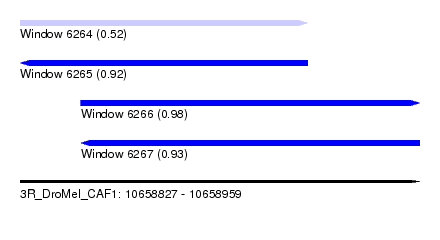
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,658,827 – 10,658,959 |
| Length | 132 |
| Max. P | 0.983986 |

| Location | 10,658,827 – 10,658,922 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -26.93 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10658827 95 + 27905053 AGUAUGUGGAUGUGAGUCGA------------------------GUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAU ...(((..(.......((((------------------------(.(((........))))))))(((((((((((((.((....)).)))))...))))))))..)..)))....... ( -31.30) >DroSec_CAF1 483 107 + 1 AGUAUGUGGAUGUGAGUCGAG------------AAGUGUUCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAU ..((((((..((((((.((..------------...))))))))..)))))).......((..(.(((((((((((((.((....)).)))))...)))))))).)..))......... ( -33.30) >DroSim_CAF1 484 119 + 1 AGUAUGUGGAUGUGAGUCGAGAUGUGGCAAUGGAAGUGUUCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAU .(((.((.((.(((..((((.(((((((..((((....))))..))))))).))))..)))))..(((((((((((((.((....)).)))))...)))))))))).)))......... ( -39.00) >consensus AGUAUGUGGAUGUGAGUCGAG____________AAGUGUUCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAU ...(((..(.......((((...............((....))....((((......))))))))(((((((((((((.((....)).)))))...))))))))..)..)))....... (-26.93 = -27.10 + 0.17)

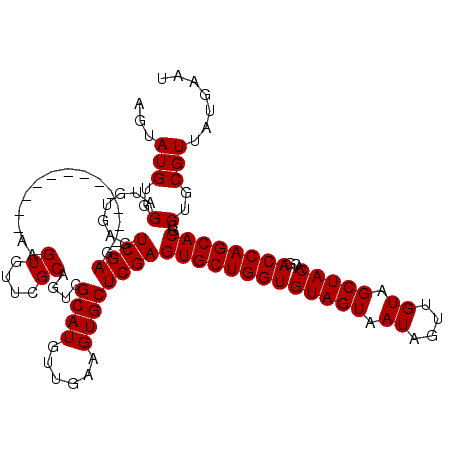
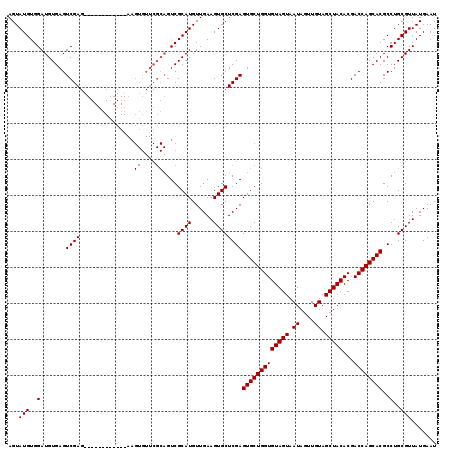
| Location | 10,658,827 – 10,658,922 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -21.03 |
| Energy contribution | -22.37 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10658827 95 - 27905053 AUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGAC------------------------UCGACUCACAUCCACAUACU ............((.((((((((.((((((.........)))).))))))))))((((((.(........).).)------------------------)))).......))....... ( -22.40) >DroSec_CAF1 483 107 - 1 AUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGAACACUU------------CUCGACUCACAUCCACAUACU ........((((((.((((((((.((((((.........)))).)))))))))).)..(((........)))..))))).......------------..................... ( -26.40) >DroSim_CAF1 484 119 - 1 AUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGAACACUUCCAUUGCCACAUCUCGACUCACAUCCACAUACU ........((((((.((((((((.((((((.........)))).)))))))))).)..(((........)))..)))))........................................ ( -26.40) >consensus AUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGAACACUU____________CUCGACUCACAUCCACAUACU ........((((((.((((((((.((((((.........)))).)))))))))).)..(((........)))..)))))........................................ (-21.03 = -22.37 + 1.33)

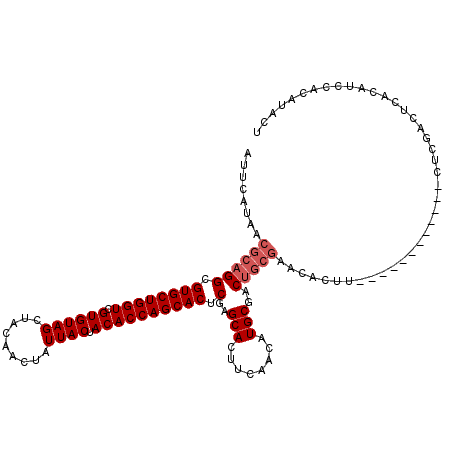
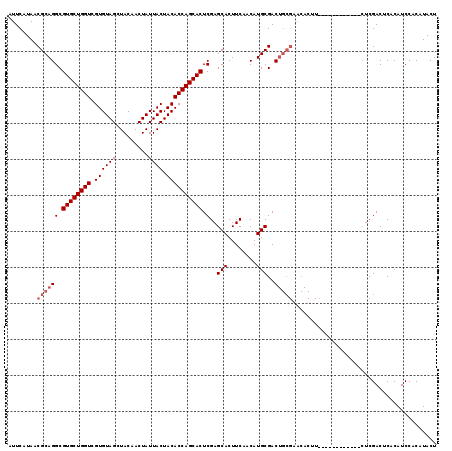
| Location | 10,658,847 – 10,658,959 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -40.87 |
| Consensus MFE | -38.43 |
| Energy contribution | -38.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10658847 112 + 27905053 -----GUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAUUAAUAAUCAUAAUAAGCCAUUAGGGUGGGCAACACGA -----...((((......))))(((.(((((((((((((.((....)).)))))...))))))))(((..((((((((........)))))))...((....)))..)))....))) ( -38.80) >DroSec_CAF1 510 117 + 1 UCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAUUAAUAAUCAUAAUAAGCCAUUAGGGUGGGCAGCACGA ..((....))........(((((...(((((((((((((.((....)).)))))...))))))))(((..((((((((........)))))))...((....)))..)))))))).. ( -41.90) >DroSim_CAF1 523 117 + 1 UCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAUUAAUAAUCAUAAUAAGCCAUUAGGGUGGGCAGCACAA ..((....))........(((((...(((((((((((((.((....)).)))))...))))))))(((..((((((((........)))))))...((....)))..)))))))).. ( -41.90) >consensus UCGCAGUCGCAUGUUGAAGUGCUCGAGUGCUGGUGUAGUAAUAGUUGUAGCUACACGACCAGCACGCCUGCGUUAUGAAUUAAUAAUCAUAAUAAGCCAUUAGGGUGGGCAGCACGA ..................(((((...(((((((((((((.((....)).)))))...))))))))(((..((((((((........)))))))...((....)))..)))))))).. (-38.43 = -38.77 + 0.33)

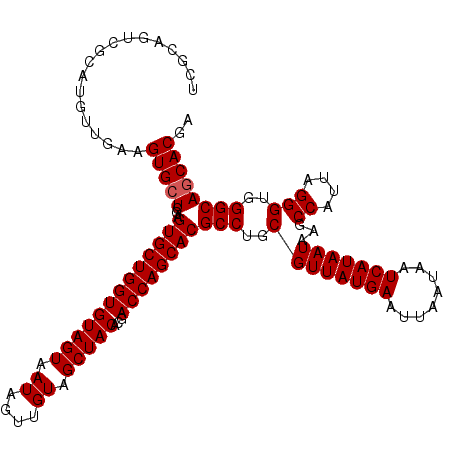
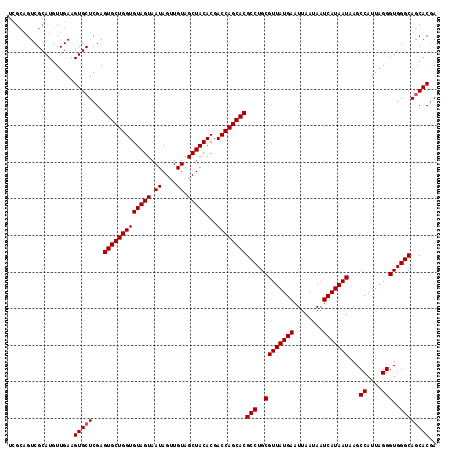
| Location | 10,658,847 – 10,658,959 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10658847 112 - 27905053 UCGUGUUGCCCACCCUAAUGGCUUAUUAUGAUUAUUAAUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGAC----- ..((((((((...((....))....((((((........))))))....)))((((((((.((((((.........)))).))))))))))...))))).............----- ( -29.20) >DroSec_CAF1 510 117 - 1 UCGUGCUGCCCACCCUAAUGGCUUAUUAUGAUUAUUAAUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGA ..((((((((...((....))....((((((........))))))....)))((((((((.((((((.........)))).))))))))))...))))).......(((....))). ( -32.50) >DroSim_CAF1 523 117 - 1 UUGUGCUGCCCACCCUAAUGGCUUAUUAUGAUUAUUAAUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGA .......(((.........)))...((((((........))))))((((((.((((((((.((((((.........)))).)))))))))).)..(((........)))..))))). ( -32.30) >consensus UCGUGCUGCCCACCCUAAUGGCUUAUUAUGAUUAUUAAUUCAUAACGCAGGCGUGCUGGUCGUGUAGCUACAACUAUUACUACACCAGCACUCGAGCACUUCAACAUGCGACUGCGA ..((((((((...((....))....((((((........))))))....)))((((((((.((((((.........)))).))))))))))...))))).................. (-30.92 = -30.70 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:13 2006