
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,654,182 – 10,654,352 |
| Length | 170 |
| Max. P | 0.988474 |

| Location | 10,654,182 – 10,654,282 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -15.41 |
| Energy contribution | -18.73 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10654182 100 + 27905053 AAUUUAAUAUAAUCAAUGCGUGGUUGCCCAGAUACAAUAA-----------AAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGA ...........(((...(((....)))...))).......-----------.................(((((((((((((.((.........)).))))))..))))))) ( -18.40) >DroSec_CAF1 3209 111 + 1 AAUUUAAUAUAAUCAAUGCGUGGUUGCCCAGGUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAGUUGUAUUUCUUCCGCCAGA ...........(((((((((((((((........))))))))..))))))).................((((((.((((((.((.........)).))))))...)))))) ( -22.10) >DroSim_CAF1 3190 111 + 1 AAUUUAAUAUAAUCAAUGCGUGGUUGCCCAGGUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGA ...........(((((((((((((((........))))))))..))))))).................(((((((((((((.((.........)).))))))..))))))) ( -25.50) >DroYak_CAF1 3217 106 + 1 AAA-----AUAAUCAAUGAGUUGUUGCCCUGAUGCAAUUAUGAAUAUUGGUAAAAUAACAAUUCAGUGUCUGGGCAGAAAUUCGUGGAAUAAUUGUAUUUCUUCGGCCAGA ...-----.......(((((((.((((((.(((((.....(((((....((......)).)))))))))).)))))).)))))))((((((....)))))).......... ( -23.60) >consensus AAUUUAAUAUAAUCAAUGCGUGGUUGCCCAGAUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGA ...........(((((((((((((((........))))))))..))))))).................(((((((((((((.((.........)).))))))..))))))) (-15.41 = -18.73 + 3.31)



| Location | 10,654,182 – 10,654,282 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.34 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -14.80 |
| Energy contribution | -16.55 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10654182 100 - 27905053 UCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUU-----------UUAUUGUAUCUGGGCAACCACGCAUUGAUUAUAUUAAAUU (((((((..((((((.((.........)).)))))))))))))...........(((...-----------(((.(((...(((....))).))).)))..)))....... ( -22.30) >DroSec_CAF1 3209 111 - 1 UCUGGCGGAAGAAAUACAACUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUACCUGGGCAACCACGCAUUGAUUAUAUUAAAUU ((((((...((((((.((.........)).)))))).)))))).....................(((.(((....(((...(((....))).))).))).)))........ ( -21.40) >DroSim_CAF1 3190 111 - 1 UCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUACCUGGGCAACCACGCAUUGAUUAUAUUAAAUU (((((((..((((((.((.........)).))))))))))))).....................(((.(((....(((...(((....))).))).))).)))........ ( -23.70) >DroYak_CAF1 3217 106 - 1 UCUGGCCGAAGAAAUACAAUUAUUCCACGAAUUUCUGCCCAGACACUGAAUUGUUAUUUUACCAAUAUUCAUAAUUGCAUCAGGGCAACAACUCAUUGAUUAU-----UUU (((((....((((((.(...........).))))))..)))))((.(((.(((((.((((..((((.......))))....)))).))))).))).)).....-----... ( -14.30) >consensus UCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUACCUGGGCAACCACGCAUUGAUUAUAUUAAAUU (((((((..((((((.((.........)).)))))))))))))...........................(((((((....(((....))).....)))))))........ (-14.80 = -16.55 + 1.75)

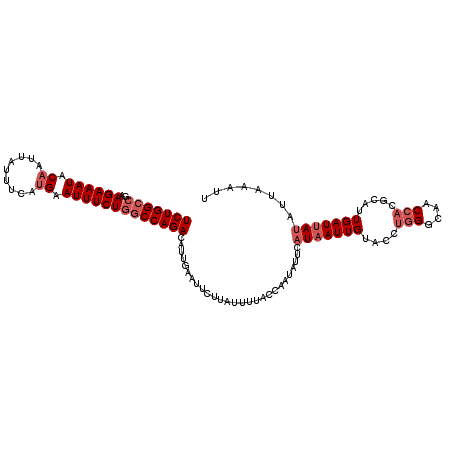

| Location | 10,654,213 – 10,654,321 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -22.68 |
| Energy contribution | -24.32 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10654213 108 + 27905053 AUACAAUAA-----------AAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGACACUGUGCUUUUGUGCCUGACUUGGGGGUGCAGUCGAAG .........-----------..........(((...((((((((((((((.((.........)).))))))..))))))))((((..(((..(.........)..)))..)))).))). ( -31.70) >DroSec_CAF1 3240 119 + 1 GUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAGUUGUAUUUCUUCCGCCAGACACUGUGCUUUUGUGUCUGACUUGGGGGUGCAGUCGAAA ......((((((((.(((((......(((........))).)))))..))))))))....(((((((..((...(.(((((((.........))))))).)..))..)))))).).... ( -30.20) >DroSim_CAF1 3221 119 + 1 GUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGACAGUGUGCUUUUGUGUCUGACUUGGGGGNNNNNNNNNNN (((((....(((((.(((......)))..))))).(((((((((((((((.((.........)).))))))..))))))))).)))))............................... ( -28.20) >DroEre_CAF1 3176 118 + 1 -UGCAAUUAAGAGUACUGGUGAGAUAACAAUUCAGUGUCUGGGCAGAAAUUCAUGGAAUAUUUGUAUAUCUUCGGCCAGACACUGUGCUUUUGUGGCUCACUUGGGGGUGCAGUCCAGC -...........(((((((((((...((((..(((((((((((((((.((((...)))).)))))....(....))))))))))).....))))..))))))....)))))........ ( -38.70) >DroYak_CAF1 3243 119 + 1 AUGCAAUUAUGAAUAUUGGUAAAAUAACAAUUCAGUGUCUGGGCAGAAAUUCGUGGAAUAAUUGUAUUUCUUCGGCCAGACACUGUGCUUUUGUGUCUGACUUGGGGGUGCAGUCCCAG .................(((......((((..((((((((((.(((((((.((.........)).))))))..).)))))))))).....)))).....)))(((((......))))). ( -31.40) >consensus AUACAAUUAUGAAUAUUGGUAAAAUAAGAAUUCAAUGUCUGGCCAGAAAUUCAUGAAAUAAUUGUAUUUCUUCGGCCAGACACUGUGCUUUUGUGUCUGACUUGGGGGUGCAGUCCAAG ...................................(((((((((((((((.((.........)).))))))..)))))))))((((((((..(.(.....).)..))))))))...... (-22.68 = -24.32 + 1.64)

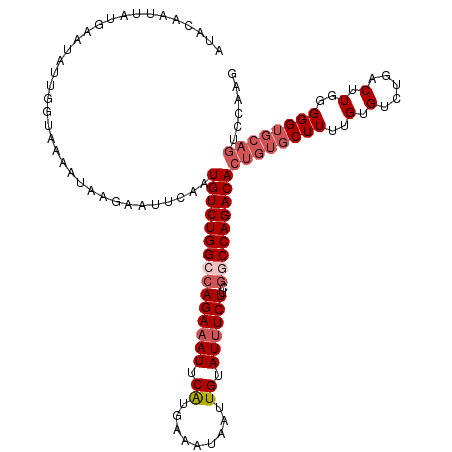
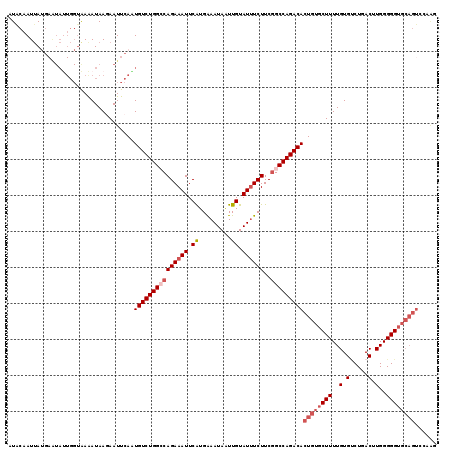
| Location | 10,654,213 – 10,654,321 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10654213 108 - 27905053 CUUCGACUGCACCCCCAAGUCAGGCACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUU-----------UUAUUGUAU ....((((.........))))..((......)).((((((((((((..((((((.((.........)).))))))))))))))))))............-----------......... ( -30.50) >DroSec_CAF1 3240 119 - 1 UUUCGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCGGAAGAAAUACAACUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUAC ....((((.........)))).............(((((((((((...((((((.((.........)).)))))).)))))))))))................................ ( -27.00) >DroSim_CAF1 3221 119 - 1 NNNNNNNNNNNCCCCCAAGUCAGACACAAAAGCACACUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUAC .........................((((........(((((((((..((((((.((.........)).))))))))))))))).(((((...............)))))...)))).. ( -22.46) >DroEre_CAF1 3176 118 - 1 GCUGGACUGCACCCCCAAGUGAGCCACAAAAGCACAGUGUCUGGCCGAAGAUAUACAAAUAUUCCAUGAAUUUCUGCCCAGACACUGAAUUGUUAUCUCACCAGUACUCUUAAUUGCA- ((((((((.........)))(((..((((.....(((((((((((.((((((((....)))))........))).).))))))))))..))))...))).))))).............- ( -28.20) >DroYak_CAF1 3243 119 - 1 CUGGGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUCCACGAAUUUCUGCCCAGACACUGAAUUGUUAUUUUACCAAUAUUCAUAAUUGCAU .((((........))))..............(((((((((((((....((((((.(...........).))))))..)))))))))).((((.........)))).........))).. ( -28.00) >consensus CUUCGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCCAGACAUUGAAUUCUUAUUUUACCAAUAUUCAUAAUUGUAC ..................................((((((((((((..((((((.((.........)).))))))))))))))))))................................ (-20.80 = -21.84 + 1.04)



| Location | 10,654,242 – 10,654,352 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -15.70 |
| Energy contribution | -18.94 |
| Covariance contribution | 3.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10654242 110 - 27905053 GUCUGCCUCUCAGCAGGAAUUUAUCCAAUUGCUUCGACUGCACCCCCAAGUCAGGCACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCC .............(((((((((((.....(((.......))).......(((((((((.........)))))))))....((((((....)))))))))))))))))... ( -27.20) >DroSec_CAF1 3280 110 - 1 GUCUGCCUCUCAGCAGGAAUUUAUCCAAUUGUUUCGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCGGAAGAAAUACAACUAUUUCAUGAAUUUCUGGCC .............(((((((((((((...(((.......))).......(((((((((.........)))))))))))).((((((....)))))).))))))))))... ( -30.10) >DroSim_CAF1 3261 110 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCCCCCAAGUCAGACACAAAAGCACACUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCC .............................................(((.((((((((...........))))))))....((((((....)))))).........))).. ( -14.50) >DroEre_CAF1 3215 110 - 1 GUCUGCCGCUCAGCGGGAAUUCAUCCAAUUGGCUGGACUGCACCCCCAAGUGAGCCACAAAAGCACAGUGUCUGGCCGAAGAUAUACAAAUAUUCCAUGAAUUUCUGCCC ............((((((((((((....(((((..(((..(((......))).((.......)).....)))..))))).(((((....)))))..)))))))))))).. ( -31.10) >DroYak_CAF1 3283 110 - 1 GUCUGCCUCUCAGCAGGAAUUUAUCCAAUUGCUGGGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUCCACGAAUUUCUGCCC (..(((.(((((((((((.....)))...))))))))..)))..)....(((((((((.........)))))))))...((((((.(...........).)))))).... ( -30.00) >consensus GUCUGCCUCUCAGCAGGAAUUUAUCCAAUUGCUUCGACUGCACCCCCAAGUCAGACACAAAAGCACAGUGUCUGGCCGAAGAAAUACAAUUAUUUCAUGAAUUUCUGGCC .............(((((((((((.........................(((((((((.........)))))))))....((((((....)))))))))))))))))... (-15.70 = -18.94 + 3.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:07 2006