| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,634,988 – 10,635,105 |
| Length | 117 |
| Max. P | 0.672561 |

| Location | 10,634,988 – 10,635,105 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -30.70 |
| Energy contribution | -31.53 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
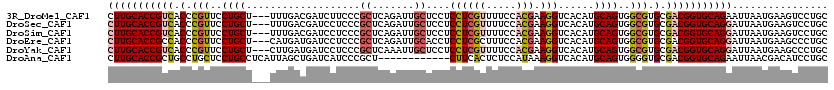
>3R_DroMel_CAF1 10634988 117 + 27905053 CUUGCACCGUCACCCGUUCCUGCU---UUUGACGAUCUUCCCGCUCAGAUUGCUCCUCCUCGUUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGAAUUAAUGAAGUCCUGC .((((((((((.(.(((..((((.---..(((((((((........)))))........((((.....))))..))))...))))..))).).))))))))))................. ( -40.00) >DroSec_CAF1 7353 117 + 1 CUUGCACCGUCACCCGUUCCUGCU---UUUGACGAUCCUCCCGCUCAGAUUGCUCCUCCUCGUUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGGAUUAAUGAAGUCCUGC ...((((((((.(.(((..((((.---..((((.........((.......))......((((.....))))..))))...))))..))).).))))))))((((((.....)))))).. ( -42.90) >DroSim_CAF1 7535 117 + 1 CUUGCACCGUCACCCGUUCCUGCU---UUUGACGAUCCUCCCGCUCAGAUUGCUCCUCCUCGUUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGGAUUAAUGAAGUCCUGC ...((((((((.(.(((..((((.---..((((.........((.......))......((((.....))))..))))...))))..))).).))))))))((((((.....)))))).. ( -42.90) >DroEre_CAF1 7408 117 + 1 CUUGCACCGCCACCCGUUCCUGCU---CAUGAUGAUCCUCCCGCUCAGAUUGCACCUCCUCGCUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGGAUUAAUGAAGCCCUGC (((((((((.(.(.(((..((((.---.(((.(((.(((..((....((..((........))..))..)).)))))))))))))..))).).).)))))))))................ ( -37.30) >DroYak_CAF1 7468 117 + 1 CUUGCACCGUCACCCGUUCCUGCU---CUUGAUGAUCCUCCCGCUCAAAUUGCUCCUCCUCGUUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGGAUUAAUGAAGCCCUGC (((((((((((.(.(((..((((.---..((.(((((.....((.......))......((((.....)))).))))))).))))..))).).)))))))))))................ ( -42.40) >DroAna_CAF1 7853 108 + 1 CUUGCACCGCUGCCUGCUCCUGCCUCAUUAGCUGAUCAUCCCGCU------------CUUCACUCUCCAUAAAGGUCACAUGCAGUGGGGUGCGACGGUGCAGAAUUAACGACAUCCUGC .(((((((((((...((....)).....))))....(((((((((------------.........((.....))........)))))))))....)))))))................. ( -29.03) >consensus CUUGCACCGUCACCCGUUCCUGCU___UUUGACGAUCCUCCCGCUCAGAUUGCUCCUCCUCGUUUUCCACGAAGGUCACAUGCAGUGGCGUGCGACGGUGCAGGAUUAAUGAAGUCCUGC (((((((((((.(.(((..((((...................((.......))....((((((.....))).)))......))))..))).).)))))))))))................ (-30.70 = -31.53 + 0.84)
| Location | 10,634,988 – 10,635,105 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -36.36 |
| Energy contribution | -35.95 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.672561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
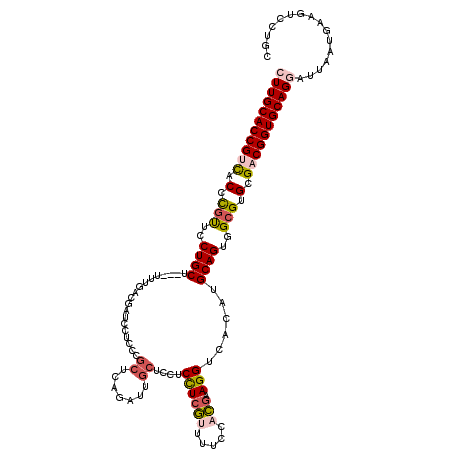
>3R_DroMel_CAF1 10634988 117 - 27905053 GCAGGACUUCAUUAAUUCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAACGAGGAGGAGCAAUCUGAGCGGGAAGAUCGUCAAA---AGCAGGAACGGGUGACGGUGCAAG ..................(((((((((((.((...((((.(((..(((((((.....)))))).)..)))...(((.(((.....))))))..---.))))...)).))))))))))).. ( -44.00) >DroSec_CAF1 7353 117 - 1 GCAGGACUUCAUUAAUCCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAACGAGGAGGAGCAAUCUGAGCGGGAGGAUCGUCAAA---AGCAGGAACGGGUGACGGUGCAAG ((((((.........))))))((((((((.((...((((.(((..(((((((.....)))))).)..)))...(((.(((.....))))))..---.))))...)).))))))))..... ( -45.20) >DroSim_CAF1 7535 117 - 1 GCAGGACUUCAUUAAUCCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAACGAGGAGGAGCAAUCUGAGCGGGAGGAUCGUCAAA---AGCAGGAACGGGUGACGGUGCAAG ((((((.........))))))((((((((.((...((((.(((..(((((((.....)))))).)..)))...(((.(((.....))))))..---.))))...)).))))))))..... ( -45.20) >DroEre_CAF1 7408 117 - 1 GCAGGGCUUCAUUAAUCCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAGCGAGGAGGUGCAAUCUGAGCGGGAGGAUCAUCAUG---AGCAGGAACGGGUGGCGGUGCAAG (((((..(.....)..)))))((((((((.((...(((((((((((((((.(....)((..(((.......)))..)).))))).))).))).---.))))...)).))))))))..... ( -42.90) >DroYak_CAF1 7468 117 - 1 GCAGGGCUUCAUUAAUCCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAACGAGGAGGAGCAAUUUGAGCGGGAGGAUCAUCAAG---AGCAGGAACGGGUGACGGUGCAAG (((((..(.....)..)))))((((((((.((...((((.(((..(((((((.....)))))).)..))).(((((..((.....)).)))))---.))))...)).))))))))..... ( -44.60) >DroAna_CAF1 7853 108 - 1 GCAGGAUGUCGUUAAUUCUGCACCGUCGCACCCCACUGCAUGUGACCUUUAUGGAGAGUGAAG------------AGCGGGAUGAUCAGCUAAUGAGGCAGGAGCAGGCAGCGGUGCAAG (((((((.......)))))))......(((((...((((.(((..(((((((.....))))))------------.............(((.....))).)..))).)))).)))))... ( -33.00) >consensus GCAGGACUUCAUUAAUCCUGCACCGUCGCACGCCACUGCAUGUGACCUUCGUGGAAAACGAGGAGGAGCAAUCUGAGCGGGAGGAUCAUCAAA___AGCAGGAACGGGUGACGGUGCAAG ((((((.........))))))((((((((.((...((((.(((..(((((((.....)))))).)..)))........((.....))..........))))...)).))))))))..... (-36.36 = -35.95 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:56 2006