
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,632,896 – 10,632,988 |
| Length | 92 |
| Max. P | 0.831450 |

| Location | 10,632,896 – 10,632,988 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10632896 92 - 27905053 AACAUGUUGGAUCGUCCGAACUUCUGGGUUGAGGGCAGCUUGGUUCUGCGUUCCAGUUUGGAUGGCAUUCGAGCCAAUUUGCCAUAGG----CCAA-------------------- ......((((.....)))).......((((...(((((.((((((((((..(((.....)))..)))...))))))).)))))...))----))..-------------------- ( -32.00) >DroVir_CAF1 5809 102 - 1 AACAUGCUAGAUCGUCCCAACUUUUGGGUGGAGGGCAGCCUUGUGCUGCGCUCGAGCCUGGAUGGCAUACGUGCCACAUUGCCAUAGG----CACAG--UACU--------CGUCA ..((.(((....((.((((.....))))))(((.(((((.....))))).))).))).))(((((..((((((((...........))----))).)--)).)--------)))). ( -36.00) >DroPse_CAF1 6447 108 - 1 AACAUGCUGGAUCGACCCAAUUUCUGGGUGGAGGGCAGCUUGGUGCUGCGCUCCAGUCUGGACGGCAUUCGAGCCACGUUGCCAUAGG----CCCCG--CC--CCCGCUCCCGCAC ....(((.(((.(((((((.....)))))((((.(((((.....))))).)))).(((((...((((..((.....)).)))).))))----)....--..--..)).))).))). ( -44.30) >DroGri_CAF1 6145 107 - 1 AAUAUGCUCGAUCGUCCCAACUUUUGGGUGGAGGGCAGUCUGGUGUUGCGCUCCAGUUUGGAUGGCAUACGUACCACAUUGCCAUAGG----CAUAGAGUUGC----CUU-CCACA ................(((.....)))((((((((((((((((((.((((..(((.......)))....)))).)))..((((...))----)))))).))))----)))-)))). ( -36.40) >DroWil_CAF1 30476 103 - 1 AAUAUGCUAGAUCGUCCGAAUUUUUGGGUGGAGGGCAGUCUCGUUUUACGUUCCAGUUUGGAUGGCACUCGAACCACGUUGCCAUAGAGAUCCCAA------------UGUAG-AU ......(((.(((..((((....))))..)).(((..(((((.........(((.....)))(((((..((.....)).)))))..))))))))..------------).)))-.. ( -26.20) >DroAna_CAF1 5891 92 - 1 AAUAUGUUAGAUCGACCCAACUUUUGGGUGGAGGGCAGCUUGGUGCUGCGUUCCAGUUUGGAUGGCAAUCGAGCCUCCUUGCCAUAGG----UCAA-------------------- .........((((.(((((.....)))))((((.(((((.....))))).)))).((..(((.(((......))))))..))....))----))..-------------------- ( -36.70) >consensus AACAUGCUAGAUCGUCCCAACUUUUGGGUGGAGGGCAGCCUGGUGCUGCGCUCCAGUUUGGAUGGCAUUCGAGCCACAUUGCCAUAGG____CCAA_______________CG___ ......((((((...((((.....))))(((((.(((((.....))))).)))))))))))((((((............))))))............................... (-21.97 = -22.47 + 0.50)

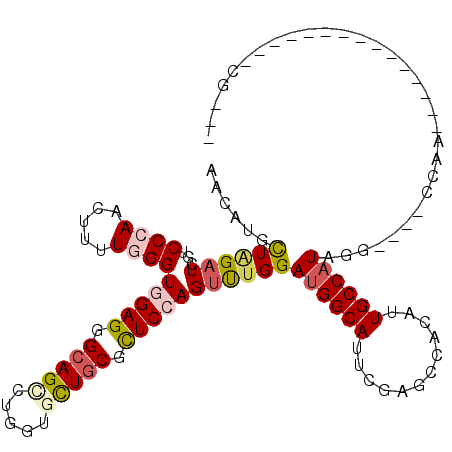
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:54 2006