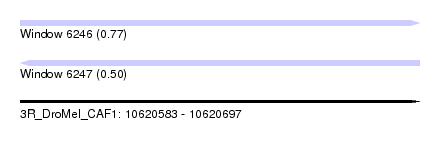
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,620,583 – 10,620,697 |
| Length | 114 |
| Max. P | 0.767515 |

| Location | 10,620,583 – 10,620,697 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.17 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.21 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
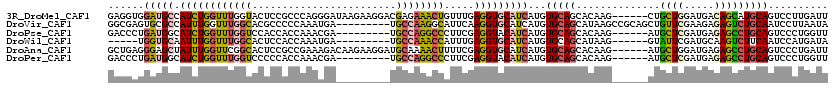
>3R_DroMel_CAF1 10620583 114 + 27905053 GAGGUGGAUGCCAUCUGGUUUGGUACUCCGCCCAGGGAUAAGAAGGACGAGAAACUGUUUGAGGUGCAUCAUGUGCAGCACAAG------CUGCUGGAUGACAGCAUGCAGUCCUUGAUU ..(((((((((((.......))))).))))))(((((((.................(((((.(.(((((...))))).).))))------)(((((.....)))))....)))))))... ( -41.90) >DroVir_CAF1 82276 111 + 1 GGCGAGUGCGCCAUUUGGUUUGGCACGCCCCCAAAUGA---------UGCCAAGGCAUUCAAGGUGCAUCAUGUGCAGCAUAAGCCGCAGCUGUUCGAAGAGAGUCUGCAAUCCUUAAUA ((((((((((((....)))((((((((........)).---------)))))).))))))..(.(((((...))))).)....)))((((((.((....)).)).))))........... ( -36.50) >DroPse_CAF1 101319 105 + 1 GACCCUGAUGGCAUCUGGUUUGGUCCACCACCAAACGA---------UGCCAGGCCCUUCGAGGUACAUCAUGUGCAGCACAAG------AUGCUCGAUGAGAGCCUGCAGUCCCUGGUU ((((..(((((((((..(((((((.....)))))))))---------))))((((.(((((((...((((.((((...)))).)------))))))))..)).))))...)))...)))) ( -39.60) >DroWil_CAF1 74263 100 + 1 -----UGGUGCAAUUUGGUUUGGCACUCCACCAAAUGA---------UGCCAAACCAUUUGAGGUGCAUCAUGUGCAGCAUAAG------GUAUUCGAUGCAAGUCUUCAAUCCAUGAUA -----(((.......((((((((((.((........))---------)))))))))).((((((((((((..((((........------))))..))))))...)))))).)))..... ( -32.10) >DroAna_CAF1 65283 114 + 1 GCUGAGGGAGCUAUUUGGUUCGGCACUCCGCCGAAAGACAAGAAGGAUGCAAAACUUUUCGAGGUGCAUCAUGUGCAGCACAAG------AUGCUGGAUGAGAGCCUGCAGUCCCUGAUU ((((.(((..((.((((.((((((.....))))))...))))...((((((...(((...))).))))))..((.(((((....------.))))).)).))..))).))))........ ( -36.70) >DroPer_CAF1 89019 105 + 1 GACCCUGAUGGCAUCUGGUUUGGUCCCCCACCAAACGA---------UGCCAGGCCCUUCGAGGUACAUCAUGUGCAGCACAAG------AUGCUCGAUGAGAGCCUGCAGUCCCUGGUU ((((..(((((((((..(((((((.....)))))))))---------))))((((.(((((((...((((.((((...)))).)------))))))))..)).))))...)))...)))) ( -39.60) >consensus GACGAGGAUGCCAUCUGGUUUGGCACUCCACCAAACGA_________UGCCAAACCAUUCGAGGUGCAUCAUGUGCAGCACAAG______AUGCUCGAUGAGAGCCUGCAGUCCCUGAUU ......(((((.((((((((((((........................)))))))).....)))))))))...(((((..............((((.....))))))))).......... (-14.06 = -14.21 + 0.14)
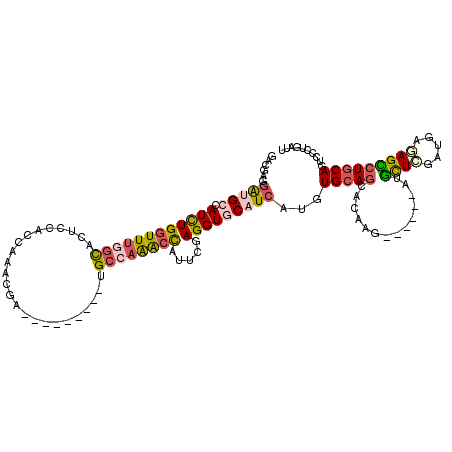
| Location | 10,620,583 – 10,620,697 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.17 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.37 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10620583 114 - 27905053 AAUCAAGGACUGCAUGCUGUCAUCCAGCAG------CUUGUGCUGCACAUGAUGCACCUCAAACAGUUUCUCGUCCUUCUUAUCCCUGGGCGGAGUACCAAACCAGAUGGCAUCCACCUC ....((((((.....(((((((((..((((------(....)))))....))))........))))).....)))))).........(((.(((...(((.......)))..))).))). ( -31.20) >DroVir_CAF1 82276 111 - 1 UAUUAAGGAUUGCAGACUCUCUUCGAACAGCUGCGGCUUAUGCUGCACAUGAUGCACCUUGAAUGCCUUGGCA---------UCAUUUGGGGGCGUGCCAAACCAAAUGGCGCACUCGCC ..((((((..((((((......))...((..((((((....))))))..)).))))))))))((((....)))---------).....(((((.((((((.......)))))).))).)) ( -36.60) >DroPse_CAF1 101319 105 - 1 AACCAGGGACUGCAGGCUCUCAUCGAGCAU------CUUGUGCUGCACAUGAUGUACCUCGAAGGGCCUGGCA---------UCGUUUGGUGGUGGACCAAACCAGAUGCCAUCAGGGUC .(((..((.....(((((((..((((((((------(.((((...)))).))))...))))))))))))((((---------(((((((((.....)))))))..)))))).))..))). ( -44.00) >DroWil_CAF1 74263 100 - 1 UAUCAUGGAUUGAAGACUUGCAUCGAAUAC------CUUAUGCUGCACAUGAUGCACCUCAAAUGGUUUGGCA---------UCAUUUGGUGGAGUGCCAAACCAAAUUGCACCA----- .....(((......((..(((((((.....------.............)))))))..))...((((((((((---------(..(.....)..))))))))))).......)))----- ( -25.87) >DroAna_CAF1 65283 114 - 1 AAUCAGGGACUGCAGGCUCUCAUCCAGCAU------CUUGUGCUGCACAUGAUGCACCUCGAAAAGUUUUGCAUCCUUCUUGUCUUUCGGCGGAGUGCCGAACCAAAUAGCUCCCUCAGC ....(((((..((((((.......((((((------...)))))).....((((((...(.....)...))))))......))))((((((.....)))))).......))))))).... ( -31.20) >DroPer_CAF1 89019 105 - 1 AACCAGGGACUGCAGGCUCUCAUCGAGCAU------CUUGUGCUGCACAUGAUGUACCUCGAAGGGCCUGGCA---------UCGUUUGGUGGGGGACCAAACCAGAUGCCAUCAGGGUC .(((..((.....(((((((..((((((((------(.((((...)))).))))...))))))))))))((((---------(((((((((.....)))))))..)))))).))..))). ( -44.00) >consensus AAUCAGGGACUGCAGGCUCUCAUCGAGCAU______CUUGUGCUGCACAUGAUGCACCUCGAAGGGCUUGGCA_________UCAUUUGGUGGAGUACCAAACCAAAUGGCACCAGCGUC ..(((((........((((.....)))).........(((.(.((((.....)))).).)))....)))))............((((((((((....)))..)))))))........... (-13.09 = -13.53 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:53 2006