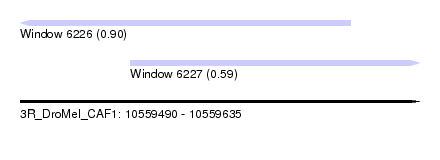
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,559,490 – 10,559,635 |
| Length | 145 |
| Max. P | 0.895401 |

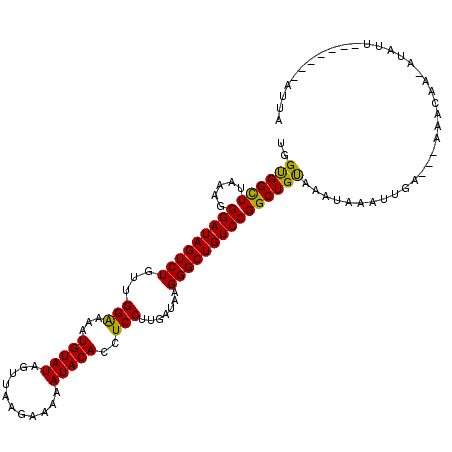
| Location | 10,559,490 – 10,559,610 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -17.97 |
| Energy contribution | -16.88 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10559490 120 - 27905053 UACAGCCGGACAGCCCUUAUCAAGGAGGUGUAUUCUUCUUAACUAUACAUUUUCCAACAGACUAUCCCUUUAAACCACCCAAAGUGGCUUUUACAACGCGCAUAUACCAUCCAAACAUCA ....((((((.((..((......((((((((((...........))))))))))....)).)).))).......((((.....))))..........).))................... ( -15.70) >DroVir_CAF1 15465 120 - 1 CACAGCCGGACAGCCCUUACCAAGGAGGUGUAUUUUUCUUAACUAUACAUUUUCCAACAGACUAUCCUUUUAAGCCACCGAAAGUGGCCUUUACAACACGCAUAUACCAUCCAAACAUUA ....((.(((.((..((......((((((((((...........))))))))))....)).)).)))......((((.(....)))))...........))................... ( -19.60) >DroGri_CAF1 31395 120 - 1 UACAGCCGGACAGCCCUUACCAAGGAGGUGUAUUUUUCUUAACUAUACAUUUCCCAACAGACUAUCCCUUUAAGCCGCCAAAAGUGGCCUUUACAACACGCAUAUACCAUCCAAACAUUA ....((.(((.((..((......((((((((((...........))))))))))....)).)).)))......(((((.....)))))...........))................... ( -21.30) >DroWil_CAF1 11155 120 - 1 UGCAGCCGGACAGCCCUUAUCAAGGAGGUGUAUUUUUCUUAACUAUACAUUUUCCAACAGACUAUCCCUUUAAACCACCAAAAGUGGCUUUUACAACACGCAUAUACCAUCCAAACAUCA (((....(((.((..((......((((((((((...........))))))))))....)).)).))).......((((.....))))............))).................. ( -16.50) >DroMoj_CAF1 8677 120 - 1 UACAGCCGGACAGCCCUUAUCAAGGAGGUGUAUUUUUCUUAACUAUACACUUUCCAACAGACUAUCCUUUUAAGCCGCCAAAAGUGGCCUUUACAACACGCAUAUACCAUCCAAACAUUA ....((.(((.((..((......((((((((((...........))))))))))....)).)).)))......(((((.....)))))...........))................... ( -21.20) >DroAna_CAF1 8039 120 - 1 UGCAGCCGGACAGCCCUUAUCAAGGAGGUGUAUUUUUCUUAACUAUACACUUUCCAACAGACUAUCCCUUUAAACCACCCAAAGUGGCUUUUACAACGCGCAUAUACCAUCCAAACAUCA (((.((.(((.((..((......((((((((((...........))))))))))....)).)).))).......((((.....))))..........))))).................. ( -20.00) >consensus UACAGCCGGACAGCCCUUAUCAAGGAGGUGUAUUUUUCUUAACUAUACAUUUUCCAACAGACUAUCCCUUUAAACCACCAAAAGUGGCCUUUACAACACGCAUAUACCAUCCAAACAUCA ....((.(((.((..((......((((((((((...........))))))))))....)).)).)))....(((((((.....)))).)))........))................... (-17.97 = -16.88 + -1.08)

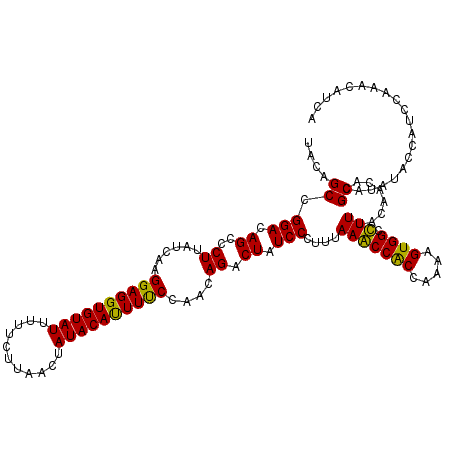
| Location | 10,559,530 – 10,559,635 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10559530 105 + 27905053 GGGUGGUUUAAAGGGAUAGUCUGUUGGAAAAUGUAUAGUUAAGAAGAAUACACCUCCUUGAUAAGGGCUGUCCGGCUGUAAGAAAAUUGA---UAACAA-AUACU-------AUUA .((((((......((((((((((((....))).....((((((.((.......)).))))))..))))))))).............(((.---...)))-..)))-------))). ( -19.80) >DroVir_CAF1 15505 112 + 1 CGGUGGCUUAAAAGGAUAGUCUGUUGGAAAAUGUAUAGUUAAGAAAAAUACACCUCCUUGGUAAGGGCUGUCCGGCUGUGAAUAUGUUUA---AAACAA-AAAUUAACACUAAUUA .(((((((.....(((((((((...(((...(((((...........)))))..))).......))))))))))))........(((...---..))).-.......))))..... ( -21.40) >DroGri_CAF1 31435 110 + 1 UGGCGGCUUAAAGGGAUAGUCUGUUGGGAAAUGUAUAGUUAAGAAAAAUACACCUCCUUGGUAAGGGCUGUCCGGCUGUAAAUAG--UCCAUCAAAUAC-AUAUA---ACCAGUUA ..((((((.....(((((((((...((((..(((((...........)))))..))))......)))))))))))))))......--............-.....---........ ( -26.40) >DroWil_CAF1 11195 102 + 1 UGGUGGUUUAAAGGGAUAGUCUGUUGGAAAAUGUAUAGUUAAGAAAAAUACACCUCCUUGAUAAGGGCUGUCCGGCUGCAAUGAAAC-------AAUAAAUUGAA-------UUGA ..(..(((.....(((((((((...(((...(((((...........)))))..))).......))))))))))))..).......(-------(((.......)-------))). ( -20.40) >DroMoj_CAF1 8717 106 + 1 UGGCGGCUUAAAAGGAUAGUCUGUUGGAAAGUGUAUAGUUAAGAAAAAUACACCUCCUUGAUAAGGGCUGUCCGGCUGUAAAUAA--UAC---AAGCAA-AUAUUU----CAAUUA ..((((((.....(((((((((...(((..((((((...........)))))).))).......)))))))))))))))......--...---......-......----...... ( -25.90) >DroAna_CAF1 8079 102 + 1 GGGUGGUUUAAAGGGAUAGUCUGUUGGAAAGUGUAUAGUUAAGAAAAAUACACCUCCUUGAUAAGGGCUGUCCGGCUGCAAAG--AU-GA---AAGAUA-CUCAU-------AUUA (((((.((((...(((((((((...(((..((((((...........)))))).))).......)))))))))..((....))--.)-))---)...))-)))..-------.... ( -25.00) >consensus UGGUGGCUUAAAGGGAUAGUCUGUUGGAAAAUGUAUAGUUAAGAAAAAUACACCUCCUUGAUAAGGGCUGUCCGGCUGUAAAUAAAUUGA___AAACAA_AUAUU_______AUUA ..((((((.....(((((((((...(((...(((((...........)))))..))).......)))))))))))))))..................................... (-20.50 = -19.67 + -0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:33 2006