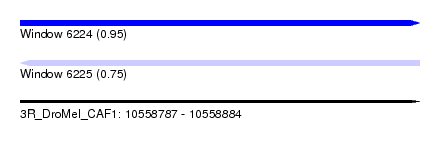
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,558,787 – 10,558,884 |
| Length | 97 |
| Max. P | 0.945960 |

| Location | 10,558,787 – 10,558,884 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Mean single sequence MFE | -41.22 |
| Consensus MFE | -27.75 |
| Energy contribution | -26.86 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10558787 97 + 27905053 -----------------UUUGCCGCUGCCGUUGCUCUUGGUACCGCCGCCAAAUUUGCUGCUCCUAAUGCUAAUGCCGCU---GCGAAGGCGGUAGCAGCGGCUGCUGCGGCGAAAA -----------------((((((((.(((((((((...(....)((((((...(((((.((.......((....)).)).---))))))))))))))))))))....)))))))).. ( -44.70) >DroPse_CAF1 10739 94 + 1 -----------------UGCACGACCCUCGUUCCCCUUGUGACGGCUAUUGUUUUUGUUGAUUUGAAUGCU---GCCACUCAGGCGGAGGCGGCGGCAGCGGCAG---GGGCGCAAU -----------------((((((.....)))..(((((((..((((..........)))).......((((---(((.(((.....)))..)))))))...))))---))).))).. ( -32.30) >DroSim_CAF1 7940 97 + 1 -----------------UUUGCCGCUGCCGUUGCUCUUGGUACCGCCGCCAAAUUUGCUGCUCCUAAUGCUAAUGCCGCU---GCGAAGGCGGUAGCAGCGGCUGCUGCGGCGAAAA -----------------((((((((.(((((((((...(....)((((((...(((((.((.......((....)).)).---))))))))))))))))))))....)))))))).. ( -44.70) >DroEre_CAF1 16980 114 + 1 ACUCCGAUCCGCUGACGUUUGCCGCUGCUGUUGCUCUUGGUACCGCCGCCAAAUUUGCUGCUCCUAAUGCUAAUGCCGCU---GCGAAGGCGGUAGCAGCGGCUGCUACGGCGAAGA .........(((((..((..((((((((((..((.(((.(((.(((.((.......)).)).......((....)).).)---)).)))))..)))))))))).))..))))).... ( -44.10) >DroYak_CAF1 17108 111 + 1 ---CCGGUCCGCUGGCGUUUGCCGCCGCUGUUGCUCUUGGUACCGCCGCCAAAUUUGCUGCUCCUAAUGCUAAUGCCGCU---GCGAAGGCGGUAGCAGCGGCUGCAACGGCGAAGA ---.......((.(((....)))))(((((((((....(....)(((((..........((.......))...(((..((---((....))))..)))))))).))))))))).... ( -44.70) >DroPer_CAF1 10731 94 + 1 -----------------UGCACGACCCUCGUUCCCCUUGUGGCGGCUAUUGUUUUUGUUGAUUUGAAUGCU---GCCACUCAGGCGGAGGCGGCGGCAGCGGCAG---GGGCGCAAU -----------------(((....((((..(((((((.((((((((....(((......)))......)))---)))))..))).))))((.((....)).))))---))..))).. ( -36.80) >consensus _________________UUUGCCGCCGCCGUUGCUCUUGGUACCGCCGCCAAAUUUGCUGCUCCUAAUGCUAAUGCCGCU___GCGAAGGCGGUAGCAGCGGCUGCU_CGGCGAAAA .................((((((((.(((((((((.........((((((...(((((..........((....)).......)))))))))))))))))))).))...)))))).. (-27.75 = -26.86 + -0.88)


| Location | 10,558,787 – 10,558,884 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.45 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -23.12 |
| Energy contribution | -23.07 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10558787 97 - 27905053 UUUUCGCCGCAGCAGCCGCUGCUACCGCCUUCGC---AGCGGCAUUAGCAUUAGGAGCAGCAAAUUUGGCGGCGGUACCAAGAGCAACGGCAGCGGCAAA----------------- .....(((((.((.((((((((..........))---))))))....((.((.((.((.((.......)).))....)).)).))....)).)))))...----------------- ( -37.50) >DroPse_CAF1 10739 94 - 1 AUUGCGCCC---CUGCCGCUGCCGCCGCCUCCGCCUGAGUGGC---AGCAUUCAAAUCAACAAAAACAAUAGCCGUCACAAGGGGAACGAGGGUCGUGCA----------------- ..(((((((---((.((((((((((((........)).)))))---))).......................((.......))))....))))..)))))----------------- ( -29.30) >DroSim_CAF1 7940 97 - 1 UUUUCGCCGCAGCAGCCGCUGCUACCGCCUUCGC---AGCGGCAUUAGCAUUAGGAGCAGCAAAUUUGGCGGCGGUACCAAGAGCAACGGCAGCGGCAAA----------------- .....(((((.((.((((((((..........))---))))))....((.((.((.((.((.......)).))....)).)).))....)).)))))...----------------- ( -37.50) >DroEre_CAF1 16980 114 - 1 UCUUCGCCGUAGCAGCCGCUGCUACCGCCUUCGC---AGCGGCAUUAGCAUUAGGAGCAGCAAAUUUGGCGGCGGUACCAAGAGCAACAGCAGCGGCAAACGUCAGCGGAUCGGAGU .(((((((((.((.((((((((..........))---))))))....((.((.((.((.((.......)).))....)).)).))....)).)))))...((....))....)))). ( -40.60) >DroYak_CAF1 17108 111 - 1 UCUUCGCCGUUGCAGCCGCUGCUACCGCCUUCGC---AGCGGCAUUAGCAUUAGGAGCAGCAAAUUUGGCGGCGGUACCAAGAGCAACAGCGGCGGCAAACGCCAGCGGACCGG--- ...((((((((((.((((((((..........))---))))))....((.......)).))))...))))))((((.((....((....))((((.....))))...)))))).--- ( -47.30) >DroPer_CAF1 10731 94 - 1 AUUGCGCCC---CUGCCGCUGCCGCCGCCUCCGCCUGAGUGGC---AGCAUUCAAAUCAACAAAAACAAUAGCCGCCACAAGGGGAACGAGGGUCGUGCA----------------- ..(((((((---((((.((....)).)).(((.(((..(((((---.((......................)).))))).))))))...))))..)))))----------------- ( -31.35) >consensus UUUUCGCCG_AGCAGCCGCUGCUACCGCCUUCGC___AGCGGCAUUAGCAUUAGGAGCAGCAAAUUUGGCGGCGGUACCAAGAGCAACGGCAGCGGCAAA_________________ ..............(((((((((((((((..(((....)))((....)).....................)))))).....(.....)))))))))).................... (-23.12 = -23.07 + -0.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:31 2006