
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,552,585 – 10,552,716 |
| Length | 131 |
| Max. P | 0.951869 |

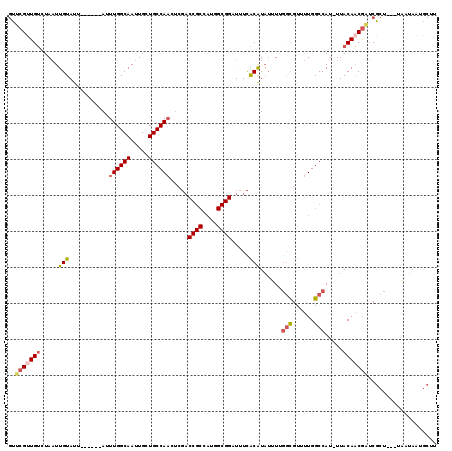
| Location | 10,552,585 – 10,552,695 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -19.13 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
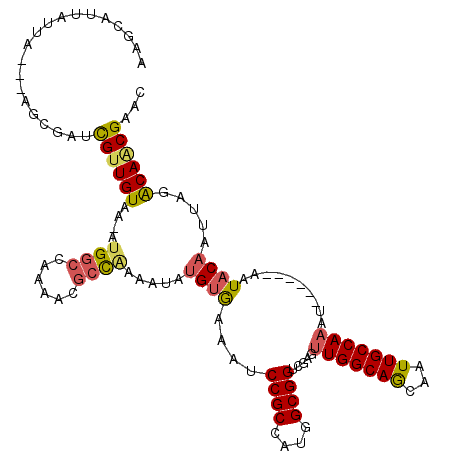
>3R_DroMel_CAF1 10552585 110 + 27905053 GUUCGUUGUCUAAUUGUAUU------AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGAUUUCACAUAUUUAAGCGUUUUGGCCAUUUUACAACGGUCGCU---UAAUAAUGCUU (..((((((..(((.((...------..((((((.....)))))).....((((....)))).......................)).)))..))))))..)((.---.......)).. ( -26.00) >DroVir_CAF1 9016 93 + 1 -UUCG-UGCCUAUUUGUAUUACGGCUAUUUGGCAAUUGUUGCCACG-----------------UUUUGCAUUGCUCGGCGUUUUGGCCAU-UUACAGCACGCACU---CGCAAA---UA -...(-(((.....((((....(((((...((((.....))))(((-----------------((..((...))..)))))..)))))..-.))))))))((...---.))...---.. ( -24.10) >DroSec_CAF1 1688 110 + 1 GUUCGUUGUCUAAUUGUAUU------AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGAUUUCACAUAUAUUGGCGUUUUGGCCAUUUUACAACGGUCGCU---UAAUAAUGCUU (..((((((.....(((...------..((((((.....)))))).....((((....)))).....))).....((((......))))....))))))..)((.---.......)).. ( -29.20) >DroSim_CAF1 1756 110 + 1 GUUCGUUGUCUAAUUGUAUU------AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGAUUUCACAUAUUUUGGCGUUUUGGCCAUUUUACAACGAUCGCU---UAAUAAUGCUU (.(((((((.....(((...------..((((((.....)))))).....((((....)))).....))).....((((......))))....))))))).)((.---.......)).. ( -30.10) >DroEre_CAF1 10836 112 + 1 GUUCGCUGUCUAAUUGUAUU------AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGAUUUCACAUAUUUUGGCGUUUUGGCCAU-UUACAACGAUCGCCUCCUAUUAACGCUC ....((.(((...(((((..------..((((((.....)))))).....((((....)))).............((((......)))).-.))))).))).))............... ( -25.80) >DroYak_CAF1 10787 112 + 1 GUUCGUUGUCUAAUUGUAUU------AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGUUUUCACGUAUUUCGCUGUUUUGGCCAA-UUACAACGAUCGCUCAAUAAUAAUGCUU (.(((((((..(((((....------..((((((.....))))))...((((((....))))))............(((.....))))))-))))))))).)((...........)).. ( -28.70) >consensus GUUCGUUGUCUAAUUGUAUU______AUUUGGCAAUUGCUGCCAACUCGACCGCCAUGGCGGAUUUCACAUAUUUUGGCGUUUUGGCCAU_UUACAACGAUCGCU___UAAUAAUGCUU ..(((((((.....(((...........((((((.....)))))).....((((....)))).....)))......(((......))).....)))))))................... (-19.13 = -20.05 + 0.92)


| Location | 10,552,585 – 10,552,695 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -20.83 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10552585 110 - 27905053 AAGCAUUAUUA---AGCGACCGUUGUAAAAUGGCCAAAACGCUUAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU------AAUACAAUUAGACAACGAAC ..(.((((((.---..((((((((.....(((((.....(((........)))......)))))))))))))...((((((...)))))))))------))).)............... ( -26.90) >DroVir_CAF1 9016 93 - 1 UA---UUUGCG---AGUGCGUGCUGUAA-AUGGCCAAAACGCCGAGCAAUGCAAAA-----------------CGUGGCAACAAUUGCCAAAUAGCCGUAAUACAAAUAGGCA-CGAA- ((---((((..---..((((.(((((..-.((((......))))............-----------------..(((((.....))))).)))))))))...))))))....-....- ( -23.80) >DroSec_CAF1 1688 110 - 1 AAGCAUUAUUA---AGCGACCGUUGUAAAAUGGCCAAAACGCCAAUAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU------AAUACAAUUAGACAACGAAC ..((.......---.))...((((((....((((......)))).....((((....((((....)))).....(((((((...)))))))..------..)))).....))))))... ( -29.00) >DroSim_CAF1 1756 110 - 1 AAGCAUUAUUA---AGCGAUCGUUGUAAAAUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU------AAUACAAUUAGACAACGAAC ..((.......---.))..(((((((....((((......)))).....((((....((((....)))).....(((((((...)))))))..------..)))).....))))))).. ( -30.60) >DroEre_CAF1 10836 112 - 1 GAGCGUUAAUAGGAGGCGAUCGUUGUAA-AUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU------AAUACAAUUAGACAGCGAAC ..((.((.....)).))..(((((((..-.((((......)))).....((((....((((....)))).....(((((((...)))))))..------..)))).....))))))).. ( -32.00) >DroYak_CAF1 10787 112 - 1 AAGCAUUAUUAUUGAGCGAUCGUUGUAA-UUGGCCAAAACAGCGAAAUACGUGAAAACCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU------AAUACAAUUAGACAACGAAC ..((.(((....)))))..(((((((((-((((((((..(.(((.....)))....(((((....))))).)..))))).((....)).....------....)))))..))))))).. ( -30.50) >consensus AAGCAUUAUUA___AGCGAUCGUUGUAA_AUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCAGCAAUUGCCAAAU______AAUACAAUUAGACAACGAAC ....................((((((....((((......)))).....((((....((((....)))).....(((((((...)))))))..........)))).....))))))... (-20.83 = -21.20 + 0.37)

| Location | 10,552,618 – 10,552,716 |
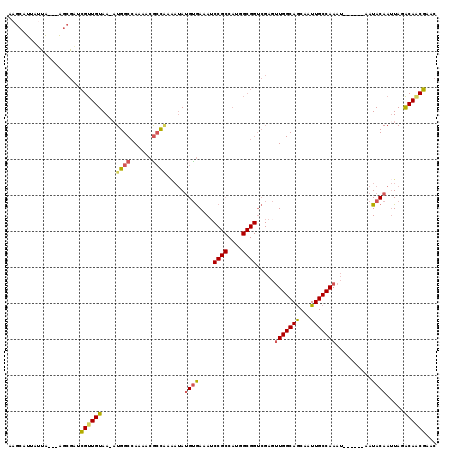
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 80.99 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.88 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
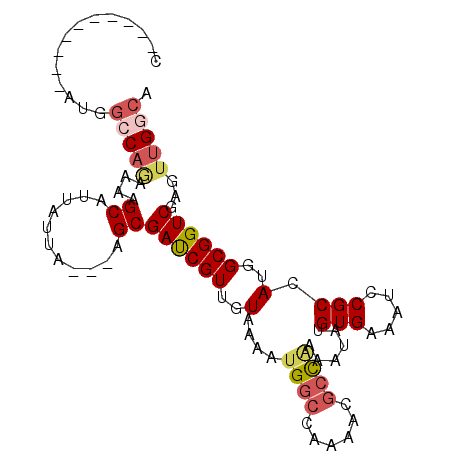
>3R_DroMel_CAF1 10552618 98 - 27905053 CGUAUGUAUGUAUGGCCAGAAAAGCAUUAUUA---AGCGACCGUUGUAAAAUGGCCAAAACGCUUAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCA (((((....)))))(((((....((((..(((---((((.(((((....)))))......)))))))..)))).....((((....)))).....))))). ( -28.40) >DroSec_CAF1 1721 90 - 1 CAA--------AUGGCCAGAGAAGCAUUAUUA---AGCGACCGUUGUAAAAUGGCCAAAACGCCAAUAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCA ...--------...(((((....((.......---.))((((((..(....((((......))))......(((.....))).)..))))))...))))). ( -26.90) >DroSim_CAF1 1789 90 - 1 CGA--------AUGGCCAGAAAAGCAUUAUUA---AGCGAUCGUUGUAAAAUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCA ...--------.((((((.....(((......---.........)))....))))))....(((((......(((...((((....)))))))..))))). ( -25.06) >DroEre_CAF1 10869 90 - 1 ----------UACGGGCACAAGAGCGUUAAUAGGAGGCGAUCGUUGUAA-AUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCA ----------....(((((((((.((((.......)))).)).))))..-...))).....(((((......(((...((((....)))))))..))))). ( -23.40) >DroYak_CAF1 10820 84 - 1 ----------------CCAAAAAGCAUUAUUAUUGAGCGAUCGUUGUAA-UUGGCCAAAACAGCGAAAUACGUGAAAACCGCCAUGGCGGUCGAGUUGGCA ----------------((((...((.(((....)))))((((((..(..-..(((.......(((.....))).......))))..))))))...)))).. ( -19.24) >consensus C__________AUGGCCAGAAAAGCAUUAUUA___AGCGAUCGUUGUAAAAUGGCCAAAACGCCAAAAUAUGUGAAAUCCGCCAUGGCGGUCGAGUUGGCA ..............(((((....((...........))((((((..(....((((......))))......(((.....))).)..))))))...))))). (-16.92 = -17.88 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:28 2006