
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,549,946 – 10,550,106 |
| Length | 160 |
| Max. P | 0.965817 |

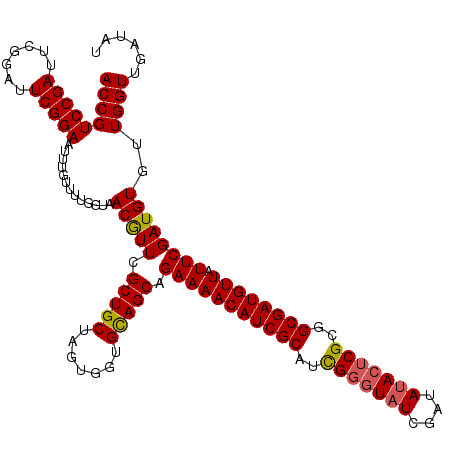
| Location | 10,549,946 – 10,550,066 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -36.09 |
| Energy contribution | -36.43 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10549946 120 + 27905053 ACCGUCCGAUUCGGAUUCGGAAUUUGUUUUGGUAACAUUCGCUGCUAGUGGUGUAGCAGAAAACAUCGCAUUGCAUAUCGAUACACUCGCGGCGAUGUUAUUCGAUGUAUUGGUUGAUAU (((((((((.......))))).......((((((((((.((((((..(((((((........)))))))((((.....))))......)))))))))))).)))).....))))...... ( -33.40) >DroSec_CAF1 24801 120 + 1 ACCGUCCGAUUCGGAUUCGGAAUUUGUUUUGGUAACGUUCGCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAU (((((((((.......))))).............(((((.(((((.......))))).(((((((((((..(((((((....)))))))..)))))))).))))))))..))))...... ( -40.20) >DroSim_CAF1 25141 120 + 1 ACCGUCCGAUUCGGAUUCGGAAUUUGUUUUGGUAACGUUCGCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAU (((((((((.......))))).............(((((.(((((.......))))).(((((((((((..(((((((....)))))))..)))))))).))))))))..))))...... ( -40.20) >consensus ACCGUCCGAUUCGGAUUCGGAAUUUGUUUUGGUAACGUUCGCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAU (((((((((.......))))).............(((((.(((((.......))))).(((((((((((..(((((((....)))))))..)))))))).))))))))..))))...... (-36.09 = -36.43 + 0.34)


| Location | 10,549,946 – 10,550,066 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -29.53 |
| Energy contribution | -29.53 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10549946 120 - 27905053 AUAUCAACCAAUACAUCGAAUAACAUCGCCGCGAGUGUAUCGAUAUGCAAUGCGAUGUUUUCUGCUACACCACUAGCAGCGAAUGUUACCAAAACAAAUUCCGAAUCCGAAUCGGACGGU ......(((...((((((...(((((((((....)(((((....)))))..))))))))..((((((......)))))))).)))).............(((((.......))))).))) ( -29.10) >DroSec_CAF1 24801 120 - 1 AUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGCGAACGUUACCAAAACAAAUUCCGAAUCCGAAUCGGACGGU ......((((((.....(((.((((((((..((.((((....)))).))..)))))))))))((((((.......))))))...)))............(((((.......))))).))) ( -34.30) >DroSim_CAF1 25141 120 - 1 AUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGCGAACGUUACCAAAACAAAUUCCGAAUCCGAAUCGGACGGU ......((((((.....(((.((((((((..((.((((....)))).))..)))))))))))((((((.......))))))...)))............(((((.......))))).))) ( -34.30) >consensus AUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGCGAACGUUACCAAAACAAAUUCCGAAUCCGAAUCGGACGGU ......((((((.....(((.((((((((..((.((((....)))).))..)))))))))))((((((.......))))))...)))............(((((.......))))).))) (-29.53 = -29.53 + 0.01)



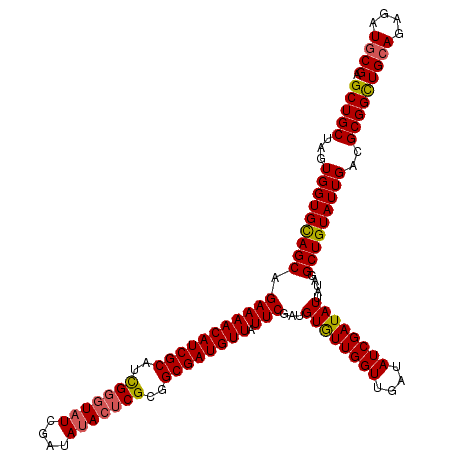
| Location | 10,549,986 – 10,550,106 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -42.13 |
| Consensus MFE | -39.25 |
| Energy contribution | -39.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10549986 120 + 27905053 GCUGCUAGUGGUGUAGCAGAAAACAUCGCAUUGCAUAUCGAUACACUCGCGGCGAUGUUAUUCGAUGUAUUGGUUGAUAUCGAUAUUAUAGGCUGUAUUGACGCGGUUGCAGAGAUGCGA .((((((......))))))......((((((((((.((((...((...(((((..(((...(((((((........)))))))....))).)))))..))...))))))))...)))))) ( -34.80) >DroSec_CAF1 24841 120 + 1 GCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAUCGAUAUUAUAGGCUGUAUUGACGCGGCUGCAGAGAUACGA (((((...(((((((((.(((((((((((..(((((((....)))))))..)))))))).)))...((((((((....)))))))).....)))))))))..)))))............. ( -44.40) >DroSim_CAF1 25181 120 + 1 GCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAUCGAUAUUAUAGGCUGUAUUGACGCGGCUGCAGAGAUGCGA (((((...(((((((((.(((((((((((..(((((((....)))))))..)))))))).)))...((((((((....)))))))).....)))))))))..)))))((((....)))). ( -47.20) >consensus GCUGCUAGUGGUGCAGCAGAAAACAUCGCAUCGGGUAUCGAUAUACUCGCGGCGAUGUUAUUCGAUGUGUUGGUUGAUAUCGAUAUUAUAGGCUGUAUUGACGCGGCUGCAGAGAUGCGA (((((...(((((((((.(((((((((((..(((((((....)))))))..)))))))).)))...((((((((....)))))))).....)))))))))..)))))((((....)))). (-39.25 = -39.70 + 0.45)


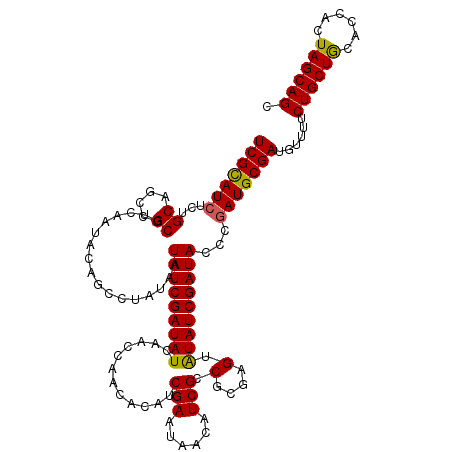
| Location | 10,549,986 – 10,550,106 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -25.03 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10549986 120 - 27905053 UCGCAUCUCUGCAACCGCGUCAAUACAGCCUAUAAUAUCGAUAUCAACCAAUACAUCGAAUAACAUCGCCGCGAGUGUAUCGAUAUGCAAUGCGAUGUUUUCUGCUACACCACUAGCAGC ((((((...(((....((.........)).....((((((((((............(((......))).(....).)))))))))))))))))))......((((((......)))))). ( -26.90) >DroSec_CAF1 24841 120 - 1 UCGUAUCUCUGCAGCCGCGUCAAUACAGCCUAUAAUAUCGAUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGC ..((((...(((....)))...))))...........(((((............)))))..((((((((..((.((((....)))).))..))))))))....(((((.......))))) ( -28.00) >DroSim_CAF1 25181 120 - 1 UCGCAUCUCUGCAGCCGCGUCAAUACAGCCUAUAAUAUCGAUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGC ..((.....((((((......................(((((............)))))..((((((((..((.((((....)))).))..))))))))....))))))......))... ( -28.40) >consensus UCGCAUCUCUGCAGCCGCGUCAAUACAGCCUAUAAUAUCGAUAUCAACCAACACAUCGAAUAACAUCGCCGCGAGUAUAUCGAUACCCGAUGCGAUGUUUUCUGCUGCACCACUAGCAGC (((((((...((....)).................(((((((((............(((......))).(....).)))))))))...)))))))......((((((......)))))). (-25.03 = -24.70 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:24 2006