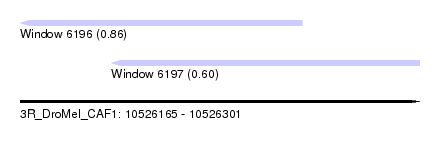
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,526,165 – 10,526,301 |
| Length | 136 |
| Max. P | 0.858482 |

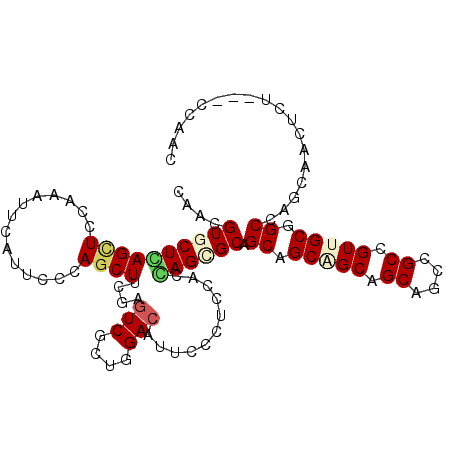
| Location | 10,526,165 – 10,526,261 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10526165 96 - 27905053 CUGGACAUUCCCUCCACCAGCGCAGCUGCAGCGGCAGCGGCCGUUGCCGCCAGCAAUGCACCAAACC---------CUGGCAACAACCGGGAGGAGUCGUCGGAU ...((((((((..((....(((..((((..((((((((....))))))))))))..)))........---------.((....))...))..))))).))).... ( -40.10) >DroPse_CAF1 1367 105 - 1 CUGGACAUUCCCUCCACGAGUGCCGCCGCAGCAGCAGCCGCCGUUGCGGCCAGCAACUCUUCGACCAGCAGCAGCACCAACAACAACAGGGAGGAGUCUUCGGAU (((((.(((((..((..((((((.((((((((.((....)).))))))))..)).))))........((....)).............))..))))).))))).. ( -41.00) >DroSec_CAF1 1507 96 - 1 CUGGACAUUCCCUCCACCAGCGCAGCUGCAGCGGCAGCGGCCGUCGCCGCAAGCAAUGCACCAAACC---------CUGGCAACAAUCGGGAGGAGUCGUCGGAU ...((((((((..((......(((..(((.(((((.((....)).)))))..))).)))........---------.((....))...))..))))).))).... ( -35.10) >DroEre_CAF1 1292 96 - 1 CUGGACAUUCCCUCCACCAGCGCAGCUGCGGCGGCAGCGGCCGUUGCCGCCAGCAAUGCUCCAAGCA---------GUGGCAACAACCGGGAGGAGUCGUCGGAU ...((((((((..((......(((..((((((((((((....))))))))).))).)))........---------.((....))...))..))))).))).... ( -42.80) >DroAna_CAF1 2802 93 - 1 CUGGACCUGCCCUCCACCAGUGCCGCAGCAGCUGCUGUCGCCGUGGCUGCCAGUAGCUCCGCGAGCA------------ACAGCAUUCGGGAGGAAUCUUCGGAC (((((..(..(((((.(.(((((.(((((....))))).(((((((..((.....)).))))).)).------------...))))).)))))).)..))))).. ( -35.20) >DroPer_CAF1 3031 105 - 1 CUGGACAUUCCCUCCACGAGUGCCGCCGCAGCAGCAGCCGCCGUUGCGGCCAGCAACUCUUCGACGAGCAGCAGCACCAACAACAACAGGGAGGAGUCUUCGGAU (((((.(((((..((..((((((.((((((((.((....)).))))))))..)).))))........((....)).............))..))))).))))).. ( -41.00) >consensus CUGGACAUUCCCUCCACCAGCGCAGCUGCAGCGGCAGCCGCCGUUGCCGCCAGCAACGCUCCAAACA_________CUGACAACAACCGGGAGGAGUCGUCGGAU ((((..(((((..((....(.((.(((((....))))).)))(((((.....)))))...............................))..)))))..)))).. (-26.40 = -26.43 + 0.03)

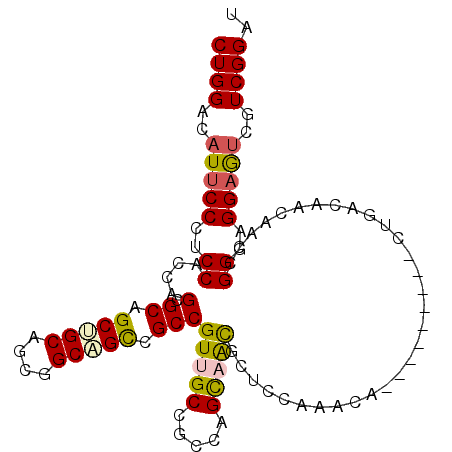
| Location | 10,526,196 – 10,526,301 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.79 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10526196 105 - 27905053 CAACGUGCUGAGCUCCAAAUUCAUUCCGAGCUUUGAGUCUCUGGACAUUCCCUCCACCAGCGCAGCUGCAGCGGCAGCGGCCGUUGCCGCCAGCAAUGCA---CCAAA ....(((((((((((............)))))....(((....)))...........)))))).((((..((((((((....))))))))))))......---..... ( -38.00) >DroPse_CAF1 1407 105 - 1 CAAUGUCCUCAGUUCGAAAUUCAUUCCCAGCUUCGAGUCGCUGGACAUUCCCUCCACGAGUGCCGCCGCAGCAGCAGCCGCCGUUGCGGCCAGCAACUCU---UCGAC .(((((((.(..((((((.............))))))..)..)))))))........((((((.((((((((.((....)).))))))))..)).)))).---..... ( -37.12) >DroEre_CAF1 1323 105 - 1 CAACGUGCUGAGCUCCAAAUUCAUUCCCAGCUUUGAGUCUCUGGACAUUCCCUCCACCAGCGCAGCUGCGGCGGCAGCGGCCGUUGCCGCCAGCAAUGCU---CCAAG ....((((((((((..............))))....(((....)))...........))))))(((((((((((((((....))))))))).)))..)))---..... ( -37.44) >DroWil_CAF1 2425 108 - 1 CAAUGUGCUUAGCUCCAAGUUCAUGCCGAGCUUCGAGUCGCUUGACAUUCCUUCCACAAGCGCAGCAGCUGCUGCCGCUGCUGUUGCGGCAAAUAACACAAACGCAAU ...((((.(((((((.((((((.....)))))).)))).(((((............)))))((((((((.((....)).))))))))......)))))))........ ( -35.30) >DroAna_CAF1 2830 105 - 1 CAACGUGCUCAGCUCCAAGUUCAUUCCCACCUUUGAAUCCCUGGACCUGCCCUCCACCAGUGCCGCAGCAGCUGCUGUCGCCGUGGCUGCCAGUAGCUCC---GCGAG ....(..(((((.((((.(((((..........)))))...)))).))).........))..)(((.(.((((((((.(((....)).).))))))))).---))).. ( -30.60) >DroPer_CAF1 3071 105 - 1 CAAUGUUCUCAGUUCGAAAUUCAUUCCCAGCUUCGAGUCGCUGGACAUUCCCUCCACGAGUGCCGCCGCAGCAGCAGCCGCCGUUGCGGCCAGCAACUCU---UCGAC .............(((((........(((((........))))).............((((((.((((((((.((....)).))))))))..)).)))))---)))). ( -36.50) >consensus CAACGUGCUCAGCUCCAAAUUCAUUCCCAGCUUCGAGUCGCUGGACAUUCCCUCCACCAGCGCAGCAGCAGCAGCAGCCGCCGUUGCGGCCAGCAACUCU___CCAAC ....((((((((((..............))))....(((....)))...........)))))).((.(((((.((....)).))))).)).................. (-18.40 = -18.79 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:12:04 2006