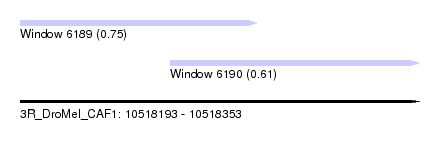
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,518,193 – 10,518,353 |
| Length | 160 |
| Max. P | 0.750422 |

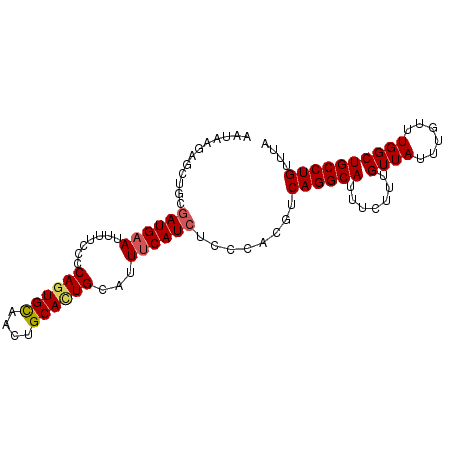
| Location | 10,518,193 – 10,518,288 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10518193 95 + 27905053 AAGAAGAGCUCCGAUGAAUUUUCCCCAGUGCAUAUGCAAUGCAUUUCAUCUCCCACGUCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA ....((.((...((((((.........((((((.....))))))))))))......((((((((.............)))))))).))))..... ( -19.32) >DroSec_CAF1 168433 95 + 1 ACCAAGAGCUGCGAUGAAUUUUCCCCAGUGCAACUGCAUUGCAUUUCAUCUCCCACGUCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA .....((((.((((((((.......((((((....))))))...))))))......((((((((.............)))))))).))..)))). ( -23.42) >DroSim_CAF1 64995 95 + 1 ACUAAGAACUGCGAUGAAUUUUCCCCAGUGCAACUGCACUGCAUUUCAUCUCCCACGUCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA .....((((.((((((((.......((((((....))))))...))))))......((((((((.............)))))))).))..)))). ( -25.72) >DroEre_CAF1 65205 95 + 1 UAUAAAAGCUGCGAUGAAUCUUGCUCAGUGUAAGUGCACUGCACUUCAUCUCCCACGGCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA .....((((...((((((...(((..(((((....)))))))).))))))......(((((.((.................)).))))).)))). ( -24.03) >DroYak_CAF1 65384 92 + 1 UAUAU---AUCCCAUGUAUUUCGCUCAGUGUAAGUGCACUGCAUUUCAUCUCCCACGGCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA .....---.................((((((....))))))...............(((((.((.................)).)))))...... ( -17.03) >consensus AAUAAGAGCUGCGAUGAAUUUUCCCCAGUGCAACUGCACUGCAUUUCAUCUCCCACGUCAGGCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUA ............((((((.......((((((....))))))...))))))........((((((.......((((......)))))))))).... (-18.23 = -18.47 + 0.24)

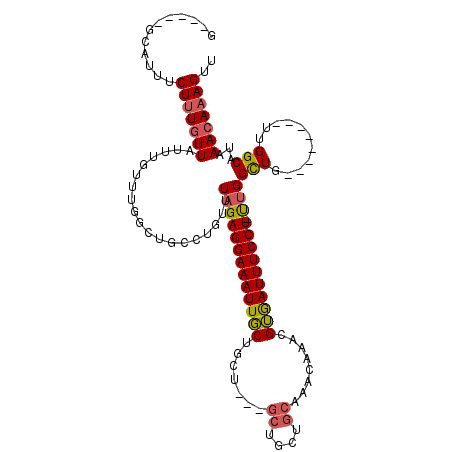
| Location | 10,518,253 – 10,518,353 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -27.99 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10518253 100 + 27905053 G-----GCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUAGAGGAAAUUGCUGCU---GCUGCUGCAAACAAACGUGAUUUCCU-UUGGCUG--------UUGGCAUAAACAAAGUU .-----......(((((((...((.(..((.(((.....(((((((((..(((.(---((....)))..))...)..)))))))-))))).)--------)..)))..))))))).. ( -28.50) >DroPse_CAF1 64126 86 + 1 GGCAUCGCAUCUCUUUCUUGUGUAUU--------UGUUUAGAGGAAAUUUCUGCU---GC-----------UACGGAAUUUCCU-CCGGUUC--------CUGGCAUAAACAAAGUC (((..((((.........))))..((--------(((((((((((((((((....---..-----------...))))))))))-)(((...--------)))...))))))))))) ( -20.70) >DroSec_CAF1 168493 101 + 1 G-----GCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUAGAGGAAAUUGCUGCU---GCUGCUGCAAACAAACGUGAUUUCCUUUUGGCUG--------UUGGCAUAAACAAAGUU .-----......(((((((...((.(..((.(((.....(((((((((..(((.(---((....)))..))...)..))))))))).))).)--------)..)))..))))))).. ( -29.70) >DroSim_CAF1 65055 101 + 1 G-----GCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUAGAGGAAAUUGCUGCU---GCUGCUGCAAACAAACGUGAUUUCCUUUUGGCUG--------UUGGCAUAAACAAAGUU .-----......(((((((...((.(..((.(((.....(((((((((..(((.(---((....)))..))...)..))))))))).))).)--------)..)))..))))))).. ( -29.70) >DroEre_CAF1 65265 100 + 1 G-----GCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUAGAGGAAAUUGCUGCU---GCUGCUGCAAACAAACGUGAUUUCCU-UUGGCUG--------CUGGCAUAAACAAAGUU .-----......(((((((...((.(..((.(((.....(((((((((..(((.(---((....)))..))...)..)))))))-))))).)--------)..)))..))))))).. ( -30.00) >DroAna_CAF1 54242 99 + 1 G-----GCACUGCUCUGUUGGUUG------------UUUAGAGGAAAUUGCUGCUGGUGCUGGUGCAAACAAACGUGAUUUCCU-CUGGCUGGCUGCUGGAUGGCGUAAACAAAGUU .-----.....(((.((((((((.------------.(((((((((((..(......(((....))).......)..)))))))-))))..))))(((....)))...)))).))). ( -29.32) >consensus G_____GCAUUUCUUUGUUAUUUGUUUGGCUGCCUGUUUAGAGGAAAUUGCUGCU___GCUGCUGCAAACAAACGUGAUUUCCU_UUGGCUG________UUGGCAUAAACAAAGUU ............(((((((...................(((((((((((((.......((....))........)))))))))).)))(((...........)))...))))))).. (-14.90 = -14.96 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:57 2006