| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,516,299 – 10,516,413 |
| Length | 114 |
| Max. P | 0.797353 |

| Location | 10,516,299 – 10,516,413 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.27 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
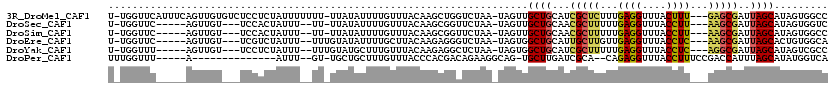
>3R_DroMel_CAF1 10516299 114 + 27905053 U-UGGUUCAUUUCAGUUGUGUCUCCUCUAUUUUUUU-UUAUAUUUUGUUUACAAGCUGGUCUAA-UAGUUGCUGCAUCGCUCUUUGAGGUUUACUUU---GAGCGAUUAGCAUAGUGGCC .-.(((.(((((((((((((.......(((......-..))).......))).)))))).....-....(((((.(((((((...(((.....))).---)))))))))))).))))))) ( -25.84) >DroSec_CAF1 166458 104 + 1 U-UGGUUC-----AGUUGU---UCCACUAUUU--UU-UUAUAUUUUGUUUACAAGCGGUUCUAA-UAGUUGCUGCAACGCUUUUUGAGGUUUACCUU---AAGCGAUUAGCAUAGUGGUC .-......-----......---.(((((((..--..-.................(((((.....-.....)))))..((((...(((((....))))---)))))......))))))).. ( -23.00) >DroSim_CAF1 62760 104 + 1 U-UGGUUC-----AGUUGU---UCCACUAUUU--UU-UUAUAUUUUGUUUACAAGCGGUUCUAA-UAGUUGCUGCAACGCUUUUUGAGGUUUACCUU---AAGCGAUUAGCAUAGUGGCC .-......-----......---.(((((((..--..-.................(((((.....-.....)))))..((((...(((((....))))---)))))......))))))).. ( -22.80) >DroEre_CAF1 63318 105 + 1 U-UGGUUC-----AGUUGU---UCGUCUAUUU--UUUGUAUAUUUUGCUUACAAGAGGGUCUAA-UAGUGGCUGCAUUGCUUGUUGAGGUUUACCUC---AAGCGAUUAGCACUGUGGCA .-......-----......---..(((..(((--((((((.........))))))))).....(-(((((.(((.((((((...(((((....))))---))))))))))))))))))). ( -28.20) >DroYak_CAF1 63404 105 + 1 U-UGGUUU-----AGUUGU---UCCUCUAUUU--UUUGUAUGCUUUGUUUACAAGAGGCUCUAA-UAGUGGCUGCAUCGCUUUUUGAGGUUUACCUC---AGGCGAUUAGCAUAGUCGCC .-....((-----((....---.(((((....--..((((.((...)).)))))))))..))))-..((((((((((((((...(((((....))))---)))))))..))..)))))). ( -28.00) >DroPer_CAF1 62118 95 + 1 UUUGGUUU-----A--------------AUUU--GU-UGCUGCUUUGUUUACCCACGACAGAAGGCAG-UGCUUGAUCGCA--CAGAGGUUUACCUUUCCGACCAUUUAGCAUAUGGUCA ...((...-----.--------------....--..-((((..((((((.......)))))).))))(-(((......)))--)(((((....)))))))((((((.......)))))). ( -25.50) >consensus U_UGGUUC_____AGUUGU___UCCUCUAUUU__UU_UUAUAUUUUGUUUACAAGCGGCUCUAA_UAGUUGCUGCAUCGCUUUUUGAGGUUUACCUU___AAGCGAUUAGCAUAGUGGCC ......................................................................((((...(((((...((((....))))...)))))..))))......... (-12.84 = -12.87 + 0.03)

| Location | 10,516,299 – 10,516,413 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.27 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -7.55 |
| Energy contribution | -8.22 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10516299 114 - 27905053 GGCCACUAUGCUAAUCGCUC---AAAGUAAACCUCAAAGAGCGAUGCAGCAACUA-UUAGACCAGCUUGUAAACAAAAUAUAA-AAAAAAAUAGAGGAGACACAACUGAAAUGAACCA-A ((......((((.(((((((---...............)))))))..))))....-......(((.((((........(((..-......)))..(....)))))))).......)).-. ( -20.46) >DroSec_CAF1 166458 104 - 1 GACCACUAUGCUAAUCGCUU---AAGGUAAACCUCAAAAAGCGUUGCAGCAACUA-UUAGAACCGCUUGUAAACAAAAUAUAA-AA--AAAUAGUGGA---ACAACU-----GAACCA-A ..((((((((((...(((((---.(((....)))....)))))....))).....-..........(((....))).......-..--..))))))).---......-----......-. ( -19.60) >DroSim_CAF1 62760 104 - 1 GGCCACUAUGCUAAUCGCUU---AAGGUAAACCUCAAAAAGCGUUGCAGCAACUA-UUAGAACCGCUUGUAAACAAAAUAUAA-AA--AAAUAGUGGA---ACAACU-----GAACCA-A ..((((((((((...(((((---.(((....)))....)))))....))).....-..........(((....))).......-..--..))))))).---......-----......-. ( -19.30) >DroEre_CAF1 63318 105 - 1 UGCCACAGUGCUAAUCGCUU---GAGGUAAACCUCAACAAGCAAUGCAGCCACUA-UUAGACCCUCUUGUAAGCAAAAUAUACAAA--AAAUAGACGA---ACAACU-----GAACCA-A .....(((((((.((.((((---((((....)))))...))).))..)))..(((-((.(.....)(((((.........))))).--.)))))....---...)))-----).....-. ( -17.20) >DroYak_CAF1 63404 105 - 1 GGCGACUAUGCUAAUCGCCU---GAGGUAAACCUCAAAAAGCGAUGCAGCCACUA-UUAGAGCCUCUUGUAAACAAAGCAUACAAA--AAAUAGAGGA---ACAACU-----AAACCA-A (((.....(((..(((((.(---((((....)))))....)))))))))))....-((((..(((((((((.........))))..--....))))).---....))-----))....-. ( -23.90) >DroPer_CAF1 62118 95 - 1 UGACCAUAUGCUAAAUGGUCGGAAAGGUAAACCUCUG--UGCGAUCAAGCA-CUGCCUUCUGUCGUGGGUAAACAAAGCAGCA-AC--AAAU--------------U-----AAACCAAA .((((((.......))))))((.((((((.......(--(((......)))-)))))))((((..((......))..))))..-..--....--------------.-----...))... ( -24.01) >consensus GGCCACUAUGCUAAUCGCUU___AAGGUAAACCUCAAAAAGCGAUGCAGCAACUA_UUAGAACCGCUUGUAAACAAAAUAUAA_AA__AAAUAGAGGA___ACAACU_____GAACCA_A .........(((...(((((....(((....)))....)))))....)))...................................................................... ( -7.55 = -8.22 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:54 2006