
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,511,833 – 10,512,015 |
| Length | 182 |
| Max. P | 0.989928 |

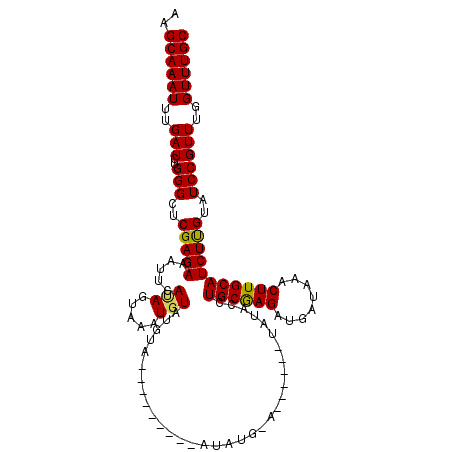
| Location | 10,511,833 – 10,511,948 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10511833 115 + 27905053 AAGCAAAUUUGAUCUGGGCUCGAGAAAUUCAUAGUAAAUGUAUGUAGAUACAUUCAUAUGUAUGUAUGUGUACCUGCGAGAUGCUAAACUUGCAUCUUGUAUCCGUUUGGUUUGC ..((((((..(((..(((......................((((((.((((((....)))))).))))))....((((((((((.......))))))))))))))))..)))))) ( -30.00) >DroSec_CAF1 160442 93 + 1 AAGCAAAUUUGAUCUGGGCUCGAGAAAUUCAUAGUAAAUUUAUGUA----------------------UAUACCUGCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGC ..((((((..(((..(((..(((((....(((((.....)))))..----------------------......((((((........)))))))))))..))))))..)))))) ( -22.10) >DroSim_CAF1 58314 91 + 1 AAGCAAAUUUGAUCUGGGCUCGAGAAAUUCAUAGUAAAUGU--GUG----------------------UAUACCUGCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGC ..((((((..(((..(((..(((((.((.((((.....)))--).)----------------------).....((((((........)))))))))))..))))))..)))))) ( -21.60) >DroEre_CAF1 58671 107 + 1 AAGCAAAUUUGAUCUGGGCUCGAGAAACUCACAGUAAAUGUAUGUC--------CAUAUGUACGUGUAUGUACCUGCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUGGGUUUGC ..(((((((..((..(((..(((((.....(((....((((((((.--------...))))))))...)))...((((((........)))))))))))..)))))..))))))) ( -29.60) >DroYak_CAF1 58809 101 + 1 AAGCAAAUUUGAUCUGGGCUCGAGAAACUCACAGUAAAUGUAUCUA--------UAUAUGCAC------AUACCUGCGAGAUGAUAAACUUGCAUCUCGUAUCCGUUUGGUUUGC ..((((((..(((..(((..(((((........(((..(((((...--------...))))).------.))).((((((........)))))))))))..))))))..)))))) ( -25.90) >consensus AAGCAAAUUUGAUCUGGGCUCGAGAAAUUCAUAGUAAAUGUAUGUA_________AUAUG_A______UAUACCUGCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGC ..((((((..(((..(((..(((((.....(((.....))).................................((((((........)))))))))))..))))))..)))))) (-20.44 = -20.08 + -0.36)

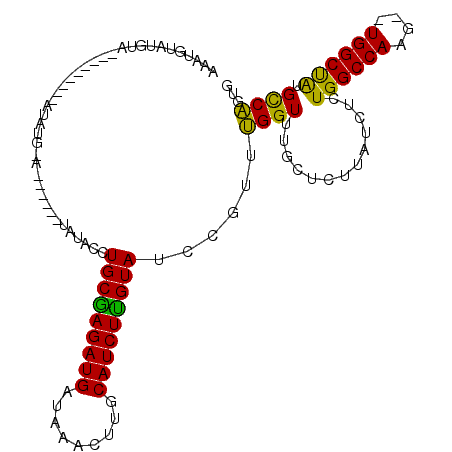
| Location | 10,511,868 – 10,511,979 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -18.06 |
| Energy contribution | -16.86 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10511868 111 + 27905053 AAAUGUAUGUAGAUACAUUCAUAUGUAUGUAUGUGUACCUGCGAGAUGCUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAU--UGGCUAUGCCAAUG (((((((((((.((((((....)))))).))))))....((((((((((.......))))))))))..)))))((((.............)))).((--((((...)))))). ( -30.82) >DroSec_CAF1 160477 91 + 1 AAAUUUAUGUA----------------------UAUACCUGCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAGUAUGGCUAUGCCAUUG ........(((----------------------...((((((((((((.........))))))))).......))).))).........((((((....))))))........ ( -18.31) >DroSim_CAF1 58349 89 + 1 AAAUGU--GUG----------------------UAUACCUGCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAGUAUGGCUACGUCAUUG .(((((--(((----------------------((.((((((((((((.........))))))))).......))).)))..........(((((....)))))))).)))). ( -21.41) >DroEre_CAF1 58706 103 + 1 AAAUGUAUGUC--------CAUAUGUACGUGUAUGUACCUGCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUGGGUUUGCUCUUAUCUCUGGCCAAG--UGGCCAUGCCUGUG ....(((..((--------((...(((((....))))).(((((((((.........))))))))).....))))..))).........(((((...--.)))))........ ( -26.30) >DroYak_CAF1 58844 97 + 1 AAAUGUAUCUA--------UAUAUGCAC------AUACCUGCGAGAUGAUAAACUUGCAUCUCGUAUCCGUUUGGUUUGCUCUUAUCGCUGGCCAAG--UGGCUGUGCCAGUG ...........--------.....((((------(....(((((((((.........))))))))).(((((((((..((.......))..))))))--))).)))))..... ( -29.30) >consensus AAAUGUAUGUA_________AUAUG_A______UAUACCUGCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAG__UGGCUAUGCCAGUG .......................................(((((((((.........)))))))))......((((.............((((((....)))))).))))... (-18.06 = -16.86 + -1.20)


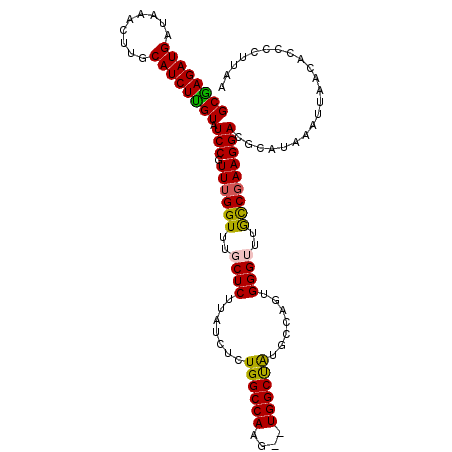
| Location | 10,511,908 – 10,512,015 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -27.58 |
| Energy contribution | -27.06 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10511908 107 + 27905053 GCGAGAUGCUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAU--UGGCUAUGCCAAUGGGUUUGCCGAAGGACGCAUAAAUUAACACCCCUUAA (((((((((.......))))))))).(((.((((((..((((.......(((((...--.))))).......))))..)))))))))...................... ( -34.44) >DroSec_CAF1 160495 109 + 1 GCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAGUAUGGCUAUGCCAUUGGGUUUGCCGAAGGACGCAUAAAUUAACACCCCUUAA (((((........)))))....((..(((.((((((..((((.......((((((....)))))).......))))..)))))))))..)).................. ( -31.54) >DroSim_CAF1 58365 109 + 1 GCAAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAGUAUGGCUACGUCAUUGGGUUUGCCGAAGGACGCAUAAAUUAACACCCCUUAA (((((........)))))....((..(((.((((((..((((.......((((((....)))))).......))))..)))))))))..)).................. ( -31.54) >DroEre_CAF1 58738 107 + 1 GCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUGGGUUUGCUCUUAUCUCUGGCCAAG--UGGCCAUGCCUGUGGGAUAGUCGAAGGACGCAUAAAUUAACACCCCUUAA ((((((((.........))))))))....(((((...(((...(((((((((((...--.))))).......))))))(((....))))))....)))))......... ( -31.81) >DroYak_CAF1 58870 107 + 1 GCGAGAUGAUAAACUUGCAUCUCGUAUCCGUUUGGUUUGCUCUUAUCGCUGGCCAAG--UGGCUGUGCCAGUGGGAUAAUCGAAGGACGCAUAAAUUAACACCCCUUAA ((((((((.........)))))))).(((.(((((((...(((...(((((((((..--....)).)))))))))).))))))))))...................... ( -30.70) >consensus GCGAGAUGAUAAACUUGCAUCUUGUAUCCGUUUGGUUUGCUCUUAUCUCUGGCCAAG__UGGCUAUGCCAGUGGGUUUGCCGAAGGACGCAUAAAUUAACACCCCUUAA ((((((((.........)))))))).(((.((((((..((((.......((((((....)))))).......))))..)))))))))...................... (-27.58 = -27.06 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:52 2006