
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,503,454 – 10,503,562 |
| Length | 108 |
| Max. P | 0.842583 |

| Location | 10,503,454 – 10,503,562 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -16.66 |
| Energy contribution | -18.22 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
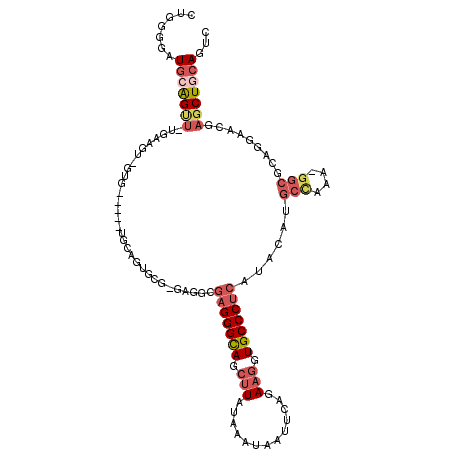
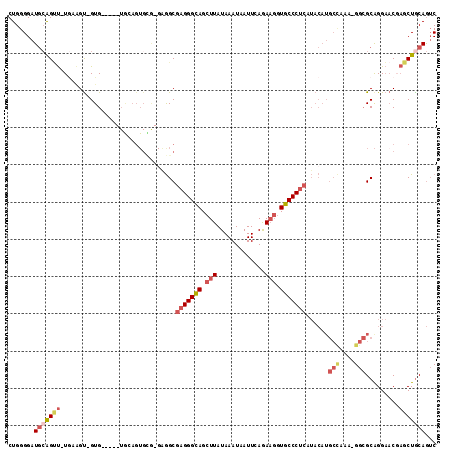
>3R_DroMel_CAF1 10503454 108 + 27905053 GACUGCAGCUCGUUCCUGCGCCAUUUGGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCCUC-CGCACUGCA-----CAC-ACUUGAAAACUGCAUCCCCAG ((.(((((.(((....((((((....)))......(((((((.(((.(((.....))).))).)))))))((...-.))...)))-----...-...)))...)))))))..... ( -29.70) >DroSec_CAF1 152140 106 + 1 GACUGCAGCUCGUUCCUGCGCC-UUUGGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCCUC-CGCACUGCA-----CAC-ACUUCA-AACUGCAUCCCCAG ...(((((..((.....(((((-...)))......(((((((.(((.(((.....))).))).)))))))))...-))..)))))-----...-......-.............. ( -27.20) >DroSim_CAF1 50064 106 + 1 GACUGCAGCUCGUUCCUGCGCC-UUUGGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCCUC-CACACUGCA-----CAC-ACUUCA-AACUGCAUCCCCAG ((.(((((...((...((((((-...)))......(((((((.(((.(((.....))).))).))))))).....-......)))-----...-))....-..)))))))..... ( -26.20) >DroEre_CAF1 50486 97 + 1 GACUGCAGCUAGUUCCCGCGCC-UUUAGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCC---------------UCCAC-ACAACA-AGCUGCAUCCCCAG ((.(((((((.(((...((...-....))......(((((((.(((.(((.....))).))).)))))))...---------------.....-..))).-)))))))))..... ( -30.20) >DroYak_CAF1 50230 112 + 1 GACUGUAGCUAGUUCCUGCGCC-UUUGGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCCCCACACACUCUACACUCCAC-ACUACA-AACUGCAUCCCCAG ((.(((((...((....(((((-...)))......(((((((.(((.(((.....))).))).))))))))).....))...))))).))...-......-.............. ( -25.10) >DroAna_CAF1 42607 91 + 1 GCUUGCAGU-CG-ACCUGCUCC-UG------------GGGCAUAGUUUGAAUUAUUUAUAAGCUACCCUCGCU-C-UCCACU-CA-----CACCGCCUUA-GCCCCAACCCCCAG ....((((.-..-..))))...-((------------((((.(((((((((....)).))))))).....((.-.-......-..-----....))....-))))))........ ( -18.94) >consensus GACUGCAGCUCGUUCCUGCGCC_UUUGGCAUGUAUGAGGGCACCUUCUGAAUUAUUUAUAAGCUGCCCUCGCCUC_CACACUGCA_____CAC_ACUUCA_AACUGCAUCCCCAG ...(((((...........((......))......(((((((.(((.(((.....))).))).))))))).................................)))))....... (-16.66 = -18.22 + 1.56)



| Location | 10,503,454 – 10,503,562 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -20.58 |
| Energy contribution | -22.09 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10503454 108 - 27905053 CUGGGGAUGCAGUUUUCAAGU-GUG-----UGCAGUGCG-GAGGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCCAAAUGGCGCAGGAACGAGCUGCAGUC .((..(((...)))..))...-...-----((((((.((-...(((((((((.(((.............))).)))))))......(((....))))).....)).))))))... ( -37.22) >DroSec_CAF1 152140 106 - 1 CUGGGGAUGCAGUU-UGAAGU-GUG-----UGCAGUGCG-GAGGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCCAAA-GGCGCAGGAACGAGCUGCAGUC .......(((((((-((...(-.((-----(((...(((-.....(((((((.(((.............))).))))))).....)))....-.))))).)..)))))))))... ( -38.92) >DroSim_CAF1 50064 106 - 1 CUGGGGAUGCAGUU-UGAAGU-GUG-----UGCAGUGUG-GAGGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCCAAA-GGCGCAGGAACGAGCUGCAGUC .......(((((((-((...(-.((-----(((....((-(.(..(((((((.(((.............))).)))))))...)...)))..-.))))).)..)))))))))... ( -39.82) >DroEre_CAF1 50486 97 - 1 CUGGGGAUGCAGCU-UGUUGU-GUGGA---------------GGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCUAAA-GGCGCGGGAACUAGCUGCAGUC .......(((((((-..((.(-((...---------------...(((((((.(((.............))).)))))))......((....-.))))).))...)))))))... ( -33.12) >DroYak_CAF1 50230 112 - 1 CUGGGGAUGCAGUU-UGUAGU-GUGGAGUGUAGAGUGUGUGGGGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCCAAA-GGCGCAGGAACUAGCUACAGUC (((......))).(-((((((-....(((.....((((.(..((((((((((.(((.............))).)))))))......)))..)-.))))....))).))))))).. ( -34.72) >DroAna_CAF1 42607 91 - 1 CUGGGGGUUGGGGC-UAAGGCGGUG-----UG-AGUGGA-G-AGCGAGGGUAGCUUAUAAAUAAUUCAAACUAUGCCC------------CA-GGAGCAGGU-CG-ACUGCAAGC ((((((......((-....))(((.-----((-(((...-(-(((.......)))).......))))).)))...)))------------))-)..((((..-..-.)))).... ( -27.80) >consensus CUGGGGAUGCAGUU_UGAAGU_GUG_____UGCAGUGCG_GAGGCGAGGGCAGCUUAUAAAUAAUUCAGAAGGUGCCCUCAUACAUGCCAAA_GGCGCAGGAACGAGCUGCAGUC .......(((((((...............................(((((((.(((.............))).)))))))......(((....))).........)))))))... (-20.58 = -22.09 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:43 2006