| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,494,037 – 10,494,140 |
| Length | 103 |
| Max. P | 0.853906 |

| Location | 10,494,037 – 10,494,140 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
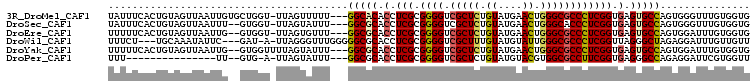
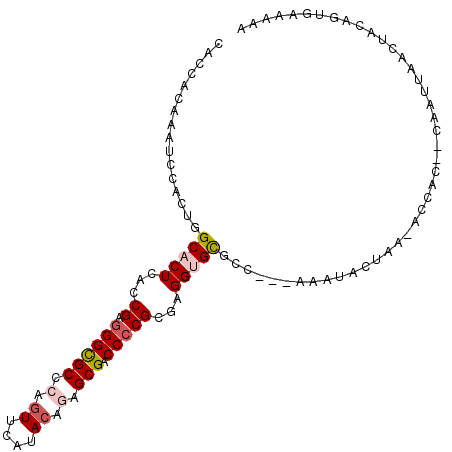
>3R_DroMel_CAF1 10494037 103 + 27905053 UAUUUCACUGUAGUUAAUUGUGCUGGU-UUAGUUUUU---GGCACACCUCGCGGGGUCGCUCUGUAUGAACUGGGCGCCCUCGGUGAGUGCCAGUGGGUUUGUGGUG .....(((..(((.....(((((..(.-.......).---.))))).(((((((((.(((((.((....)).))))).))))(((....))).))))).)))..))) ( -37.00) >DroSec_CAF1 40134 101 + 1 UAUUUCACUGUAGUUAAUUU--GUGGU-UUAGUAUUU---GGCGCACCUCGCGGGGUCGCUCUGUAUGAACUGGGCACCCUCGGUGAGUGCCAGUGGGUUUGUGGUG ...(((((((..(((((.((--(....-.)))...))---)))((((.(((((((((.((((.((....)).)))))))))..)))))))))))))))......... ( -31.30) >DroEre_CAF1 40892 101 + 1 UUUUUCACUGUAGUUAAUUG--GUGGU-UUAGUGUUU---GGCGCACCUCGCGGGGUCGCUCUGUAUGAACUGGGCGCCCUCGGUGAGUGCCAGUGGAUUUGUGGUG .....(((..(((...((((--((...-...(((...---..)))..(((((((((.(((((.((....)).))))).)))).))))).))))))....)))..))) ( -35.40) >DroWil_CAF1 38015 99 + 1 UUUCU---UGCAAAUAUUC---GAU-A-UUAGGGUUUGGGGGCGCACCUCGCGGGGUCGCUUUGUAUGUAUUGGGCGCCCUCGGUUAGGGCUAGAGGAUUUGUUGUU .....---.((((((.(((---...-.-.........((((((((((((....)))).((.......)).....))))))))(((....))).))).)))))).... ( -26.90) >DroYak_CAF1 40636 102 + 1 UUUUUCACUGUAGUUAAUUG--GUGGUUUUAGUAUUU---GGCGCACCUCGCGGGGUCGCUCUGUAUGAACUGGGCGCCCUCGGUGAGUGCCAGUGGAUUUGUGGUG ...(((((((..(((((.((--.((....)).)).))---)))((((.((((((((.(((((.((....)).))))).)))).)))))))))))))))......... ( -34.90) >DroPer_CAF1 35945 85 + 1 UUU---------------UU--GUG-A-UUAGUAUUU---GGCGCACCUCGCGGGGUCGCUCUGUAUGUACGUGGCGCCUUCGGUGAGGGCCAGAGGAUUCGUGGUU ...---------------(.--.((-(-......(((---(((...((((((((((.((((.(((....))).)))).)))).))))))))))))....)))..).. ( -29.70) >consensus UUUUUCACUGUAGUUAAUUG__GUGGU_UUAGUAUUU___GGCGCACCUCGCGGGGUCGCUCUGUAUGAACUGGGCGCCCUCGGUGAGUGCCAGUGGAUUUGUGGUG ........................................(((((.(.(((.((((.(((((.((....)).)))))))))))).).)))))............... (-22.42 = -22.65 + 0.23)
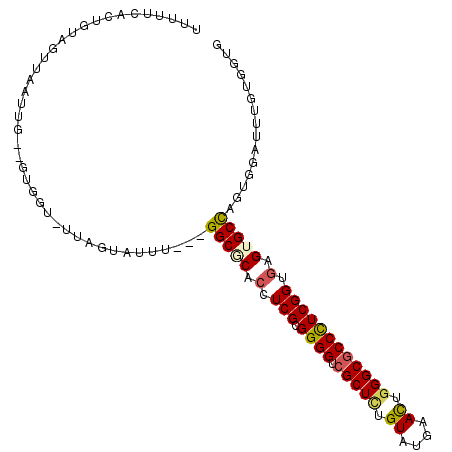
| Location | 10,494,037 – 10,494,140 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -13.99 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10494037 103 - 27905053 CACCACAAACCCACUGGCACUCACCGAGGGCGCCCAGUUCAUACAGAGCGACCCCGCGAGGUGUGCC---AAAAACUAA-ACCAGCACAAUUAACUACAGUGAAAUA ...........(((((...(((..((.((((((.(.((....)).).))).))))).))).(((((.---.........-....)))))........)))))..... ( -25.54) >DroSec_CAF1 40134 101 - 1 CACCACAAACCCACUGGCACUCACCGAGGGUGCCCAGUUCAUACAGAGCGACCCCGCGAGGUGCGCC---AAAUACUAA-ACCAC--AAAUUAACUACAGUGAAAUA ...(((......((((((((((.....))))).))))).......(.(((.(.((....)).)))))---.........-.....--............)))..... ( -24.70) >DroEre_CAF1 40892 101 - 1 CACCACAAAUCCACUGGCACUCACCGAGGGCGCCCAGUUCAUACAGAGCGACCCCGCGAGGUGCGCC---AAACACUAA-ACCAC--CAAUUAACUACAGUGAAAAA ...........(((((...(((...)))(((((((.((....))...(((....)))..)).)))))---.........-.....--..........)))))..... ( -24.90) >DroWil_CAF1 38015 99 - 1 AACAACAAAUCCUCUAGCCCUAACCGAGGGCGCCCAAUACAUACAAAGCGACCCCGCGAGGUGCGCCCCCAAACCCUAA-U-AUC---GAAUAUUUGCA---AGAAA .....(((((..((..(((((.....)))))................(((.(.((....)).)))).............-.-...---))..)))))..---..... ( -20.20) >DroYak_CAF1 40636 102 - 1 CACCACAAAUCCACUGGCACUCACCGAGGGCGCCCAGUUCAUACAGAGCGACCCCGCGAGGUGCGCC---AAAUACUAAAACCAC--CAAUUAACUACAGUGAAAAA ...........(((((...(((...)))(((((((.((....))...(((....)))..)).)))))---...............--..........)))))..... ( -24.90) >DroPer_CAF1 35945 85 - 1 AACCACGAAUCCUCUGGCCCUCACCGAAGGCGCCACGUACAUACAGAGCGACCCCGCGAGGUGCGCC---AAAUACUAA-U-CAC--AA---------------AAA .....((...(((((((((((......))).))))............(((....)))))))..))..---.........-.-...--..---------------... ( -19.70) >consensus CACCACAAAUCCACUGGCACUCACCGAGGGCGCCCAGUUCAUACAGAGCGACCCCGCGAGGUGCGCC___AAAUACUAA_ACCAC__CAAUUAACUACAGUGAAAAA ................(((((...((.((((((.(.((....)).).))).)))))...)))))........................................... (-13.99 = -14.72 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:34 2006