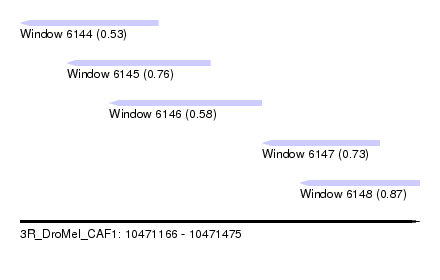
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,471,166 – 10,471,475 |
| Length | 309 |
| Max. P | 0.868023 |

| Location | 10,471,166 – 10,471,273 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.99 |
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.51 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10471166 107 - 27905053 CGAGUCGGAGCUGAUAGGCACUAACGAUGCUGGUAAGUUUCAAAACGGUGAAA-CUUUUAUGACUUCCAUAUAAUAGCUAUGUAUAUAUAGAAGGAAGCUGGACAAAU ...(((((((((....(((((....).))))....))))))....((((....-((((((((.....((((.......))))....))))))))...))))))).... ( -26.10) >DroEre_CAF1 17371 96 - 1 CGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCCA----------AUUUAUGUUACUUCCGUAUAAUAUUUAUGUAUAUAUAAAAG-GAGCUUGACA-AU .((((.((((((....(((((....).))))....)).)))).----------)))).(((((.((((.((((((((....)))).))))...)-)))..)))))-.. ( -20.10) >DroYak_CAF1 17584 94 - 1 CGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCGAAACGAUGGGAAUUUAUAUGACAUCCGU-------------AUAUACAAAAGGGUGCUUGACA-AU ((..((((((((....(((((....).))))....))))))))..))................(((((..-------------..........))))).......-.. ( -22.00) >consensus CGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUUCAAAACG_UG__AAUUUAUAUGACUUCCGUAUAAUA__UAUGUAUAUAUAAAAGGGAGCUUGACA_AU ...(((((((((....(((((....).))))....))))))......................(((((.........................)))))...))).... (-18.06 = -17.51 + -0.55)


| Location | 10,471,202 – 10,471,313 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.09 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10471202 111 - 27905053 CAUACGCCAGUUCCGAGGAUACCGGAUUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCACUAACGAUGCUGGUAAGUUU----CAAAACGGUGAAA-CUUUUAUGACUUCCAUAU ....((((...((((.(....)))))...))))(((((((((.((..(.(((....))).)..))..)))))).((((((----((......)))))-)))....)))........ ( -35.50) >DroSec_CAF1 17180 94 - 1 CAUACGCCAGUUCCGAGGAUACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUU----CAAAACGGUGGGA-C----------------- ....(((((((.(((.(....))))))).))))((((((.((..(.((((((....(((((....).))))....)))))----).)..)).)))))-)----------------- ( -35.70) >DroSim_CAF1 17914 94 - 1 CAUACGCCAGUUCCGAGGAUACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUU----CAAAACGGUGGGA-C----------------- ....(((((((.(((.(....))))))).))))((((((.((..(.((((((....(((((....).))))....)))))----).)..)).)))))-)----------------- ( -35.70) >DroEre_CAF1 17405 102 - 1 CAUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUU----CCA----------AUUUAUGUUACUUCCGUAU ......(((((.(((.(....)))))))))((((...(((((.((..(.(((....))).)..))..)))))((((((..----...----------)))))).......)))).. ( -30.30) >DroWil_CAF1 18234 99 - 1 CCUAUGCCAGUUCCGAAGACACGGGCCUGGGUGGCCUAAGUGAAUCGGAGCUGAUGGGCGCUAACGAUGCUGGUAAGUUAAUCACAAAAA------AAUUCAUAU----------- ..((((.(((((((((...((((((((.....)))))..)))..)))))))))((.(((((....).)))).))................------....)))).----------- ( -32.10) >DroYak_CAF1 17608 110 - 1 CGUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUU----CGAAACGAUGGGAAUUUAUAUGACAUCCGU-- (((((((((((.(((.(....))))))).)))).(((((.((..((((((((....(((((....).))))....)))))----)))..)).))))).....))))........-- ( -38.70) >consensus CAUACGCCAGUUCCGAGGACACCGGACUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGUUU____CAAAACGGUGGGA_CUUAUAU___________ ..(((..(((((((((.......(((((....))))).......)))))))))...(((((....).)))).)))......................................... (-23.34 = -23.09 + -0.25)



| Location | 10,471,235 – 10,471,353 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -46.88 |
| Consensus MFE | -31.38 |
| Energy contribution | -32.50 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10471235 118 - 27905053 CUUGGCCAGGACAUGUUUGGUAUUUGCGAGAACAGCUCGGCAUACGCCAGUUCCGAGGAUACCGGAUUGGGCGGUCUCAGCGAGUCGGAGCUGAUAGGCACUAACGAUGCUGGUAAGU-- ((((.((....((.((((.(.((((((((((...(((((((....)))...((((.(....)))))..))))..)))).))))))).))))))...(((((....).)))))))))).-- ( -40.20) >DroVir_CAF1 15107 118 - 1 CUGGGCCAGCACAUGUUUGGCAUCUGCGAAAACAGCUCGGCCUAUGCCAGCUCCGAGGAUACGGGCCUGGGUGGCCUCAGUGAGUCGGAGCUGAUAGGCGCAAAUGAUGCUGGUAAGU-- ((..((((((((((((((.((....))..)))).((...((((((..(((((((((...((((((((.....)))))..)))..)))))))))))))))))..))).))))))).)).-- ( -52.40) >DroGri_CAF1 23000 118 - 1 CUGAGUCAGCACAUGUUUGGCAUAUGCGAGAACAGCUCGGCCUAUGCCAGCUCCGAGGAUACGGGCCUGGGCGGUCUGAGUGAGUCGGAGCUGAUAGGCGCGAAUGAUGCGGGUAAGU-- (((.((((((...(((((.((....))..)))))))(((((((((..(((((((((..((.((((((.....)))))).))...))))))))))))))).))).)))).)))......-- ( -47.20) >DroWil_CAF1 18255 118 - 1 UUGGGUCAGGAUCUGUUUGGUGUCUGCGAUAAUAGUUCAGCCUAUGCCAGUUCCGAAGACACGGGCCUGGGUGGCCUAAGUGAAUCGGAGCUGAUGGGCGCUAACGAUGCUGGUAAGU-- .....((((.(((.(((.(((((((......((((......))))..(((((((((...((((((((.....)))))..)))..)))))))))..))))))))))))).)))).....-- ( -43.60) >DroMoj_CAF1 24886 118 - 1 CUGAGCCAGCAUAUGUUUGGCAUCUGCGAGAACAGUUCGGCCUAUGCCAGCUCCGAAGAUACGGGCCUGGGAGGUCUGAGUGAGUCGGAGCUGAUAGGCGCUAAUGAUGCUGGUAAGU-- ....((((((((....(((((.....((((.....))))((((((..(((((((((..((.((((((.....)))))).))...))))))))))))))))))))..))))))))....-- ( -55.30) >DroAna_CAF1 16452 120 - 1 UUGGGCCAAGAAAUGCUUAGUAUCUGCGAGAAUAGCUCGGCGUACGCCAGCUCAGAGGACACUGGACUGGGAGGGCUGAGCGAGUCCGAGCUGAUUGGCGCCAACGAUGCUGGUGAGUGG (((((((......(((.........)))......(((((((.(.(.((((..(((......)))..))))).).)))))))).))))))(((.((..(((.......)))..)).))).. ( -42.60) >consensus CUGGGCCAGCACAUGUUUGGCAUCUGCGAGAACAGCUCGGCCUAUGCCAGCUCCGAGGAUACGGGCCUGGGAGGUCUGAGUGAGUCGGAGCUGAUAGGCGCUAACGAUGCUGGUAAGU__ ................((((((((..((((.....))))((((((..(((((((((...(((.((((.....))))...)))..)))))))))))))))......))))))))....... (-31.38 = -32.50 + 1.12)

| Location | 10,471,353 – 10,471,444 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.49 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -25.61 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10471353 91 - 27905053 GGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA---CGCUGUUCCGGCAGUGCUGGCCAAUACGGCCGAUCUU (((((.((((...)))).))))).(((..((...........))....((((((---((((((....)))))).))))))....)))....... ( -39.30) >DroSec_CAF1 17314 91 - 1 GGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA---AGCUGUUCCGGCACCGCUGGCCAACACGGCCGAUCUG (((((.((((...)))).))))).(((..((...........))....((((((---.((.((......)).))))))))....)))....... ( -33.50) >DroSim_CAF1 18048 91 - 1 GGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA---AGCUGUUCCGGCACCGCUGGCCAACACGGCCGAUCUG (((((.((((...)))).))))).(((..((...........))....((((((---.((.((......)).))))))))....)))....... ( -33.50) >DroEre_CAF1 17547 91 - 1 GGCCUUUGGCAACGCCAUUGGUCAGCCAACCAACACAAUUACGCACAAUGGCCA---AGCUGUUCCGGCAGUGCUGGCCAACACGACCGAUCUG (((....(....))))(((((((.((................))....((((((---.(((((....)))))..))))))....)))))))... ( -29.19) >DroYak_CAF1 17758 91 - 1 GGCCUUUGGCAACGCCAUUGGUCAGCCAACCAACACAAUUACGCACAAUGGCCA---AGCCGUUCCGGCAGUGCUGGCCAACACGGCCGAUCUG ((((..((((...))))((((((((((...............((......))..---.((((...)))).).)))))))))...))))...... ( -32.80) >DroPer_CAF1 17001 79 - 1 UGCCUUCGGCAAUGCCAUUGGUCAGCCC------------ACGCACAACGGCCACCAGACGGCUCUGGC---CCUGGCCAACACGGCGAAUCUG ((((...)))).((((.(((((((((..------------..)).....(((((((....))...))))---).)))))))...))))...... ( -26.20) >consensus GGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA___AGCUGUUCCGGCAGUGCUGGCCAACACGGCCGAUCUG ((((...(((...)))...(((((((................((......))......((((...))))...))))))).....))))...... (-25.61 = -25.50 + -0.11)

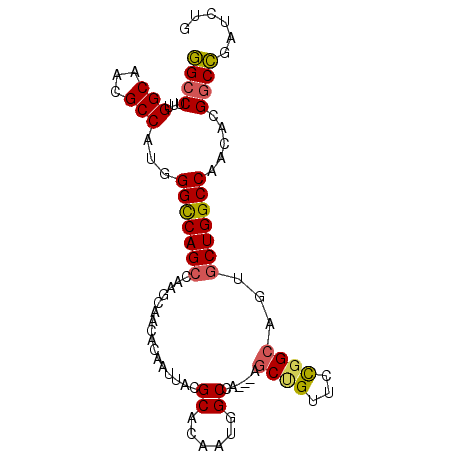
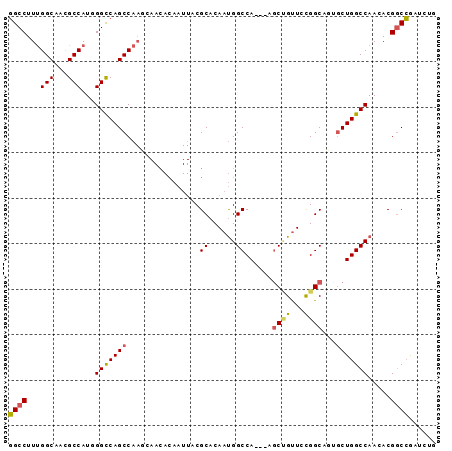
| Location | 10,471,382 – 10,471,475 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -22.01 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10471382 93 - 27905053 AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA--CGCUGUUC ........((((((.................(((((.((((...)))).))))).((((.((...........))....))))..--.)))))). ( -24.50) >DroSec_CAF1 17343 93 - 1 AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA--AGCUGUUC ........((((((((...............(((((.((((...)))).))))).((((.((...........))....)))).)--))))))). ( -26.00) >DroSim_CAF1 18077 93 - 1 AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUGGGCCAGCCAAGCAACACAAUUACGCACAAUGGCCA--AGCUGUUC ........((((((((...............(((((.((((...)))).))))).((((.((...........))....)))).)--))))))). ( -26.00) >DroEre_CAF1 17576 93 - 1 AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUUGGUCAGCCAACCAACACAAUUACGCACAAUGGCCA--AGCUGUUC ........((((((((...............((((..((((...))))..)))).((((....................)))).)--))))))). ( -21.55) >DroWil_CAF1 18410 91 - 1 GUGACAGCAUCAGUUUCCCCUCAUUUAAUACAGCCUUUGGCAAUGCCAUGGGU-GGCCAAC--AUACAAUCACGCACAACGGUAUUAAUCGGA-A ..............((((......(((((((.((((.((((...)))).))))-.......--.........((.....)))))))))..)))-) ( -16.30) >DroYak_CAF1 17787 93 - 1 AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUUGGUCAGCCAACCAACACAAUUACGCACAAUGGCCA--AGCCGUUC .............................(((((....(....)(((((((....((................)).)))))))..--.))))).. ( -17.69) >consensus AUAACAAUAACAGUUUCCCCUCAUUCAACACGGCCUUUGGCAACGCCAUGGGCCAGCCAACCAACACAAUUACGCACAAUGGCCA__AGCUGUUC ........(((((((................((((..((((...))))..)))).((((....................))))....))))))). (-17.83 = -18.33 + 0.50)

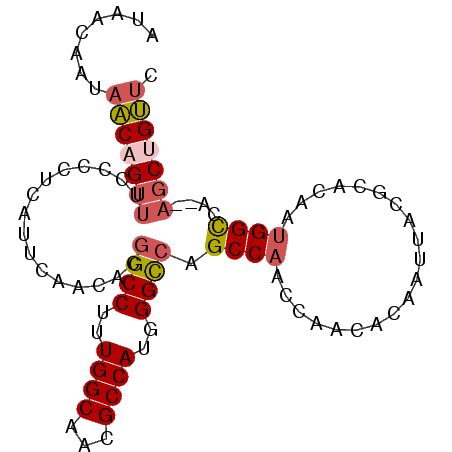
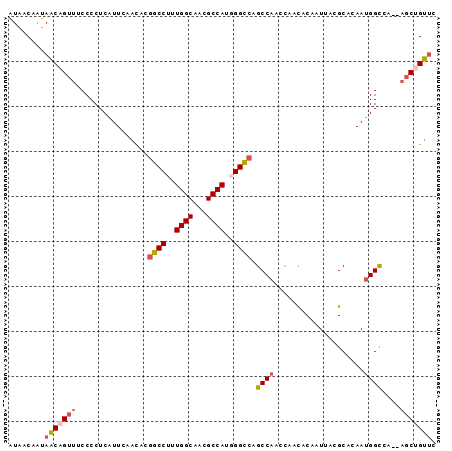
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:18 2006