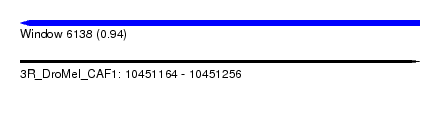
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,451,164 – 10,451,256 |
| Length | 92 |
| Max. P | 0.935547 |

| Location | 10,451,164 – 10,451,256 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
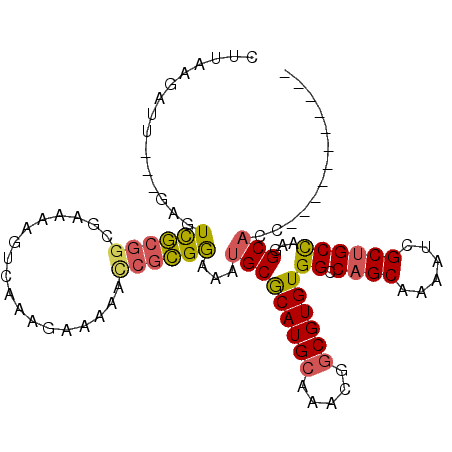
>3R_DroMel_CAF1 10451164 92 - 27905053 CUUAAAAUU---AAGUCGCGGCGAAAAGUCGAGUAAAACCCGCGGAAAUGCGCAUGCAAACGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCACC----------- .........---...(((((((((....)))........))))))...(((((((((.....))))))(((.(((......))))))...)))..----------- ( -26.60) >DroSec_CAF1 4424 92 - 1 CUUAAGAUU---GAGUCGCGGUGAAAGGUCAAAGAAAAACCGCGGAAAUGCGCAUGCAAACGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCACC----------- .........---...(((((((................)))))))...(((((((((.....))))))(((.(((......))))))...)))..----------- ( -28.39) >DroSim_CAF1 4473 92 - 1 CUUAAGAUU---GAGUCGCGGUGAAAAGUCGAAGAAAAACCGCGGAAAUGCGCAUGCAAACGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCAUC----------- .........---...(((((((................)))))))..((((((((((.....))))))(((.(((......))))))...)))).----------- ( -29.09) >DroEre_CAF1 4553 92 - 1 CUUACGAUC---GAGUCGCGGCGAAAAGUCAAGGAAAAUCCGCGGAAAUGCGCAUGCAAACGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCACC----------- .........---..((((((((((........((.....))((((.....(((((((.....))))))).)).))....)))))))...)))...----------- ( -27.90) >DroYak_CAF1 4446 92 - 1 CCUACGAUC---GAGUCGCGGCGACAAGUCAAGCAUAAUUCGCGGAAAUGCGCAUGCAAGCGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCACC----------- (((......---..((((((((.....)))..((((....((((....)))).))))..)))))...((((.(((......))))))))))....----------- ( -29.50) >DroMoj_CAF1 4176 104 - 1 AAUGUGAUAGCAAAGUUAA--UGAUAAUGCAAAUCGAAAUCGUGAAAAUGCACAUGGAAAAAACGUGUGGUCCGCAACUUUGCUGCUACUGCUGCAGCAGAAAAUA .......((((((((((..--.(((...............(((....)))((((((.......)))))))))...))))))))))...((((....))))...... ( -26.10) >consensus CUUAAGAUU___GAGUCGCGGCGAAAAGUCAAAGAAAAACCGCGGAAAUGCGCAUGCAAACGGCGUGUGGCCAGCAAAAUCGCUGCCAAGGCACC___________ ...............((((((..................))))))...(((((((((.....))))))((.((((......))))))...)))............. (-21.01 = -21.07 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:08 2006