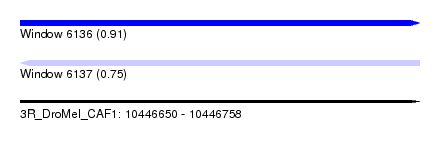
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,446,650 – 10,446,758 |
| Length | 108 |
| Max. P | 0.907206 |

| Location | 10,446,650 – 10,446,758 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -23.39 |
| Energy contribution | -22.75 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10446650 108 + 27905053 UCUUACGACCUCGACGAUCAACUAUCCACUACGAAGCAAAAAGCU-UCAACUGAUUUUUUUGCAGAGCCACAGAUCCGCUGGCUGGUCUGUGGCUUAGUACCGCUUCUA .....((....))...................(((((.(((((..-((....))..)))))((.((((((((((((.(....).)))))))))))).))...))))).. ( -31.30) >DroSec_CAF1 11451 104 + 1 UCUUACGACCUCGACGAUCAACUAUCCACUACGAAGCUAAAAGCU-UCAACUGAUUU-GUUGCAGAGCCACAGAUCCGCUGGCUGGUCUGUGGCUUAGUACCGCUU--- .....((....))...................(((((.....)))-)).........-(.(((.((((((((((((.(....).)))))))))))).))).)....--- ( -30.70) >DroSim_CAF1 11734 104 + 1 UCUUACGACCUCGACGAUCAACUAUCCACUACGAAGCUAAAAGCU-UCAACUGAUUU-GUUGCAGAGCCACAGAUCCACUGGCUGGUCUGUGGCUUAGUACCGCUU--- .....((....))...................(((((.....)))-)).........-(.(((.((((((((((.(((.....))))))))))))).))).)....--- ( -29.80) >DroEre_CAF1 5029 102 + 1 UCUUACGACC--AACGUUCAACUUUCCACUAGCAAGCGAAAAGCU-UCAGCUGAUUU-GUUGCAGAGCCACAGAGCCGCUGGCUGUUCUGUGGCUUAGUACCGCUU--- ...(((....--...................((((.((((.(((.-...)))..)))-))))).((((((((((((.(....).)))))))))))).)))......--- ( -31.40) >DroYak_CAF1 12090 104 + 1 UCUUACGACAACGCCGAUCAAUUGUCCACUACUAAGCUAAAAGCU-UCAACUGAUUU-CUUCCAGAGCCACAGAGCCGUUGACUGGUCUGUGGCUUAGUACCGCUU--- ............((.(((((.(((.........((((.....)))-)))).))))).-......((((((((((.(((.....)))))))))))))......))..--- ( -26.00) >DroPer_CAF1 4857 105 + 1 ACUUAAUUAUCCAACUAUCAACUAACGACUGCUGAGCAAAAGGCUUUCGAUUGAUUG-GUUGGCAAGUCACAGCCCUGCCCUUGGGGCUGUGACUUAGUACCGCUU--- ..........(((((((((((....(((..(((........)))..))).)))).))-))))).(((((((((((((......)))))))))))))..........--- ( -39.40) >consensus UCUUACGACCUCGACGAUCAACUAUCCACUACGAAGCUAAAAGCU_UCAACUGAUUU_GUUGCAGAGCCACAGAUCCGCUGGCUGGUCUGUGGCUUAGUACCGCUU___ .................................((((.........((....))......(((.((((((((((((........)))))))))))).)))..))))... (-23.39 = -22.75 + -0.64)

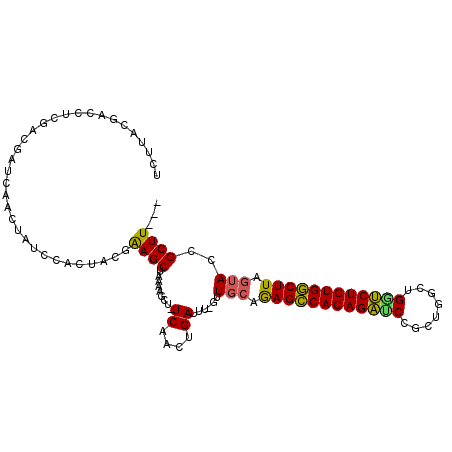
| Location | 10,446,650 – 10,446,758 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10446650 108 - 27905053 UAGAAGCGGUACUAAGCCACAGACCAGCCAGCGGAUCUGUGGCUCUGCAAAAAAAUCAGUUGA-AGCUUUUUGCUUCGUAGUGGAUAGUUGAUCGUCGAGGUCGUAAGA .....((((.....(((((((((((.......)).))))))))))))).......(((.((((-(((.....)))))).).)))......((((.....))))...... ( -33.60) >DroSec_CAF1 11451 104 - 1 ---AAGCGGUACUAAGCCACAGACCAGCCAGCGGAUCUGUGGCUCUGCAAC-AAAUCAGUUGA-AGCUUUUAGCUUCGUAGUGGAUAGUUGAUCGUCGAGGUCGUAAGA ---..((((.....(((((((((((.......)).)))))))))))))(((-...(((.((((-(((.....)))))).).)))...)))((((.....))))...... ( -33.60) >DroSim_CAF1 11734 104 - 1 ---AAGCGGUACUAAGCCACAGACCAGCCAGUGGAUCUGUGGCUCUGCAAC-AAAUCAGUUGA-AGCUUUUAGCUUCGUAGUGGAUAGUUGAUCGUCGAGGUCGUAAGA ---..((((.....((((((((((((.....))).)))))))))))))(((-...(((.((((-(((.....)))))).).)))...)))((((.....))))...... ( -34.70) >DroEre_CAF1 5029 102 - 1 ---AAGCGGUACUAAGCCACAGAACAGCCAGCGGCUCUGUGGCUCUGCAAC-AAAUCAGCUGA-AGCUUUUCGCUUGCUAGUGGAAAGUUGAACGUU--GGUCGUAAGA ---..((((.....(((((((((...(((...))))))))))))))))...-..((((((...-(((((((((((....)))))))))))....)))--)))....... ( -38.00) >DroYak_CAF1 12090 104 - 1 ---AAGCGGUACUAAGCCACAGACCAGUCAACGGCUCUGUGGCUCUGGAAG-AAAUCAGUUGA-AGCUUUUAGCUUAGUAGUGGACAAUUGAUCGGCGUUGUCGUAAGA ---...((.((((((((((((((((.......)).)))))))).(((((((-...((....))-..))))))).)))))).))((((((........))))))...... ( -33.50) >DroPer_CAF1 4857 105 - 1 ---AAGCGGUACUAAGUCACAGCCCCAAGGGCAGGGCUGUGACUUGCCAAC-CAAUCAAUCGAAAGCCUUUUGCUCAGCAGUCGUUAGUUGAUAGUUGGAUAAUUAAGU ---.........(((((((((((((........)))))))))))))(((((-..(((((((((..((..........))..)))...)))))).))))).......... ( -35.60) >consensus ___AAGCGGUACUAAGCCACAGACCAGCCAGCGGAUCUGUGGCUCUGCAAC_AAAUCAGUUGA_AGCUUUUAGCUUCGUAGUGGAUAGUUGAUCGUCGAGGUCGUAAGA .....((((.....(((((((((((.......)).)))))))))))))......((((((((...(((...........)))...))))))))................ (-20.95 = -21.23 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:11:07 2006