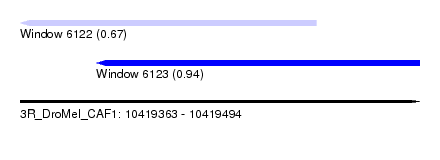
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,419,363 – 10,419,494 |
| Length | 131 |
| Max. P | 0.944796 |

| Location | 10,419,363 – 10,419,460 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
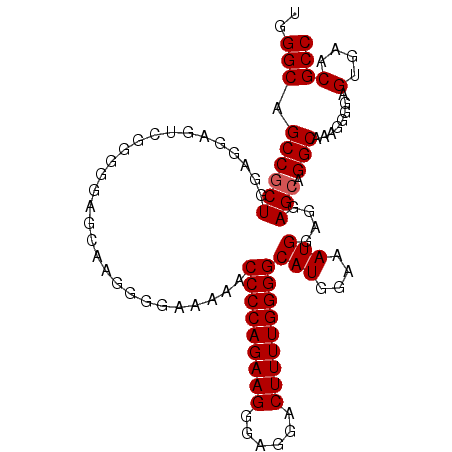
>3R_DroMel_CAF1 10419363 97 - 27905053 UGGGCAGCCACUGGAGGAGUC-----AACAGGGGGGAAAACCCCAGAAGGGAGGACUUUUGGGGCAUGAAAAUGGAGUAGCAGGCAAAGGGGAGUGAACGCC ..(((.(((.(((........-----..))).........(((((((((......)))))))))(((....)))........)))........(....)))) ( -28.00) >DroSec_CAF1 82773 102 - 1 UGGGCAGCCGCUGGAGGAGUCGGGGGAGCAAGGGGAAAACCCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAGGAGCAGGCAAAGGGGAGUGAACGCC ..(((.(((.((.((....)).))...((...........(((((((((......)))))))))(((....))).....)).)))........(....)))) ( -30.50) >DroSim_CAF1 70481 102 - 1 UGGGCAGCCGCUGGAGGAGUCGGGGGAGCAAGGGGAAAAUCCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAGGAGCGGGCAAAGGGGAGUGAACGCC ..(((.(((.((.((....)).))...((...........(((((((((......)))))))))(((....))).....)).)))........(....)))) ( -30.10) >consensus UGGGCAGCCGCUGGAGGAGUCGGGGGAGCAAGGGGAAAAACCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAGGAGCAGGCAAAGGGGAGUGAACGCC ..(((.((((((............................(((((((((......)))))))))(((....)))....))).)))........(....)))) (-26.77 = -27.10 + 0.33)


| Location | 10,419,388 – 10,419,494 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
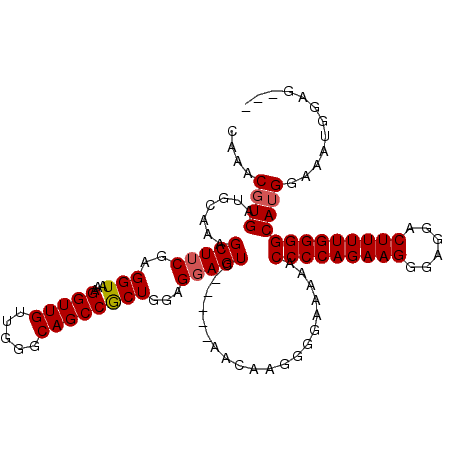
>3R_DroMel_CAF1 10419388 106 - 27905053 CAAACGUGAUGCAAAGCUGCGAGGUAAAGGUUGUUGGGCAGCCACUGGAGGAGUC-----AACAGGGGGGAAAACCCCAGAAGGGAGGACUUUUGGGGCAUGAAAAUGGAG--- ....(.((.((....(((.(..(((...(((((.....))))))))...).))))-----).)).)........(((((((((......)))))))))(((....)))...--- ( -29.10) >DroSec_CAF1 82798 111 - 1 CAAACGUGAUGCAAAGCUUCGAGGUAAAGGUUGUUGGGCAGCCGCUGGAGGAGUCGGGGGAGCAAGGGGAAAACCCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAG--- ....((((.......(((((........(((((.....))))).((.((....)).)))))))...........(((((((((......))))))))))))).........--- ( -33.90) >DroSim_CAF1 70506 111 - 1 CAAACGUGAUGCAAAGCUUCGAGGUAAAGGUUGUUGGGCAGCCGCUGGAGGAGUCGGGGGAGCAAGGGGAAAAUCCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAG--- ....((((.......(((((........(((((.....))))).((.((....)).)))))))...........(((((((((......))))))))))))).........--- ( -33.70) >DroYak_CAF1 81026 97 - 1 CAAACGUGAUGCAAAGCUUCGAGGUAAAGG-----------CCGCUGGAGGAGUU------AGUUGGGGGAAAGCCCCAGAAGGGUGGACUUUUGGGGCAGGGAAAUGGAGUGG ((..(((..(.(....((((.((.(((...-----------((......))..))------).)).))))...((((((((((......)))))))))).).)..)))...)). ( -30.10) >consensus CAAACGUGAUGCAAAGCUUCGAGGUAAAGGUUGUUGGGCAGCCGCUGGAGGAGUC_____AACAAGGGGAAAAACCCCAGAAGGGAGGACUUUUGGGGCAUGGAAAUGGAG___ ....((((.......(((((..(((...(((((.....))))))))...)))))....................(((((((((......)))))))))))))............ (-26.60 = -27.10 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:54 2006