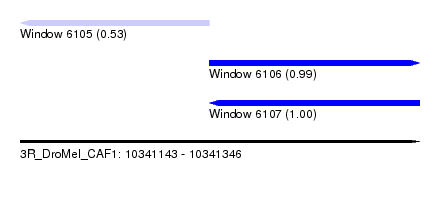
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,341,143 – 10,341,346 |
| Length | 203 |
| Max. P | 0.997230 |

| Location | 10,341,143 – 10,341,239 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -16.44 |
| Energy contribution | -16.56 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10341143 96 - 27905053 AACUCACAUUA-AAAAUCUUGUUGUGCUCCACCUGCAUCUACUACAGGAUUUGGUACUCGAUUUCGUUUGGUUUGAAUGUUUUUGUUUGUGAAACUA ...(((((...-.(((((((((.((((.......)))).....)))))))))......(((...(((((.....)))))...)))..)))))..... ( -18.20) >DroSec_CAF1 3464 96 - 1 AACUCACAUAA-AAAAUCUUGUUGUGCUCCACCUGUAUCUAUUACAGGAUUUAGUACUCGAUUUCGUUUGGUUUGAAUGUUUUUGUUUGUGAAACUU ...(((((..(-(((((.(((..(((((...((((((.....))))))....))))).))).((((.......)))).))))))...)))))..... ( -20.30) >DroSim_CAF1 3472 96 - 1 AACUCACAUAA-AAAAUCUUGUUGUGCUCAACCUGUAUCUAUUACAGGAUUUGGUACUCGAUUUCGUUUGGUUUGAAUGUUUUUGUUUGUCAAACUA ...........-.(((((.....(((((.((((((((.....)))))).)).)))))..)))))....((((((((.............)))))))) ( -18.92) >DroEre_CAF1 4013 96 - 1 AACUCACAAAA-AACAUCUUGUUGUGCUCCACCUGUACCCAUUGCAGGAUUUGGUACUCGAUUUCGUUUGGCUUGAAUGUUUUUGUUGGUGAAACUA ...((((.(((-((((((..((((((((...((((((.....))))))....))))).((....))...)))..).))))))))....))))..... ( -22.90) >DroYak_CAF1 3512 97 - 1 AACUCACAAAAAAAAAUCUUGUUGUGCUCCACCUGUAUCCAUUACAGGAUUUGGUACUCGAUUUCGUUUGGUUUGAAUGUUUUUGUUUGUGAAACUA ...(((((((.((((((.(((..(((((...((((((.....))))))....))))).))).((((.......)))).)))))).)))))))..... ( -22.30) >consensus AACUCACAUAA_AAAAUCUUGUUGUGCUCCACCUGUAUCUAUUACAGGAUUUGGUACUCGAUUUCGUUUGGUUUGAAUGUUUUUGUUUGUGAAACUA ...((((................(((((...((((((.....))))))....))))).(((...(((((.....)))))...)))...))))..... (-16.44 = -16.56 + 0.12)

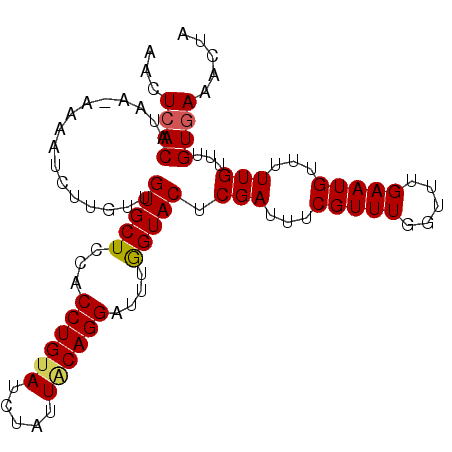
| Location | 10,341,239 – 10,341,346 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -46.05 |
| Consensus MFE | -35.16 |
| Energy contribution | -35.55 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.72 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10341239 107 + 27905053 --UUCAAA-----ACUCCAAAAUCCAAUUAGAACUCGGUCAUCCCCCCUCCG---CACUUGUGGUCAGUCUCACUGCCCGU---GGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG --......-----.........(((((((((((...(((.((...(((((((---((((...((.(((.....)))))...---))))).)))))).)).)))...)))))))))))... ( -41.30) >DroSec_CAF1 3560 107 + 1 --UUCCAA-----ACUCCAAAAUCCAAUUAGAACUCGGUCAUCCCCCCUCCG---CACUUGUGGUCAGUCUCACUGACCGU---UGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG --......-----.........(((((((((((...(((.((...(((((((---(((...(((((((.....))))))).---.)))).)))))).)).)))...)))))))))))... ( -45.30) >DroSim_CAF1 3568 107 + 1 --UUCCAA-----ACUCCAAAAUCCAAUUAGAACUCGGUCAUCCCCCCUCCG---CACUUGUGGUCAGUCUCACUGCCCGU---GGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG --......-----.........(((((((((((...(((.((...(((((((---((((...((.(((.....)))))...---))))).)))))).)).)))...)))))))))))... ( -41.30) >DroEre_CAF1 4109 107 + 1 --UUGCAA-----ACUCCAAAAUCCAAUUAGAACUCGGUCAUCCCCCCUCCG---CACUUGUGGUCAGUUUCACUGCCCGU---GGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG --......-----.........(((((((((((...(((.((...(((((((---((((...((.(((.....)))))...---))))).)))))).)).)))...)))))))))))... ( -41.30) >DroYak_CAF1 3609 107 + 1 --UUCGAA-----ACUCCAAAAUCCAAUUAGAAGUCGGUCAUCCCCCCUCCG---CACUUGUGGUCAGUUUCACUGCCCGU---GGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG --......-----.........(((((((((((...(((.((...(((((((---((((...((.(((.....)))))...---))))).)))))).)).)))...)))))))))))... ( -42.40) >DroAna_CAF1 3042 120 + 1 UACUCCCAAAUCGACGGCGAAAUCCAAUUAGAACUCGGUCAGCCCCCCUCCGGCACUCUUCGGGUCAGUCUCACUGCCCGGGGUCGUGCUGGAGGGAGUUGCCAUGUUCUAAUUGGACGG .....((...(((....)))..((((((((((((..((.((((..(((((((((((.(((((((.(((.....))))))))))..))))))))))).))))))..)))))))))))).)) ( -64.70) >consensus __UUCCAA_____ACUCCAAAAUCCAAUUAGAACUCGGUCAUCCCCCCUCCG___CACUUGUGGUCAGUCUCACUGCCCGU___GGUGCUGGAGGGAGUCGCCAUAUUCUAAUUGGACGG ......................(((((((((((...(((....(.(((((((...((((.((((.(((.....))).))))...)))).))))))).)..)))...)))))))))))... (-35.16 = -35.55 + 0.39)
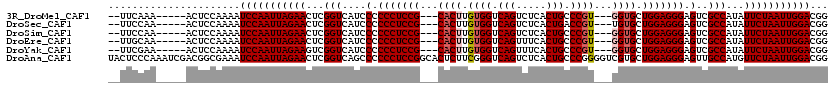

| Location | 10,341,239 – 10,341,346 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -51.55 |
| Consensus MFE | -41.06 |
| Energy contribution | -41.15 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -5.26 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10341239 107 - 27905053 CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACC---ACGGGCAGUGAGACUGACCACAAGUG---CGGAGGGGGGAUGACCGAGUUCUAAUUGGAUUUUGGAGU-----UUUGAA-- (((((((((((((((.(((((.((((((((.((((.---...(((((.....))).))....)))---)))))))))..)).))).)))))))))))))...))...-----......-- ( -49.60) >DroSec_CAF1 3560 107 - 1 CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACA---ACGGUCAGUGAGACUGACCACAAGUG---CGGAGGGGGGAUGACCGAGUUCUAAUUGGAUUUUGGAGU-----UUGGAA-- (((((((((((((((.(((((.((((((((.((((.---..((((((.....))))))....)))---)))))))))..)).))).)))))))))))))...))...-----......-- ( -53.10) >DroSim_CAF1 3568 107 - 1 CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACC---ACGGGCAGUGAGACUGACCACAAGUG---CGGAGGGGGGAUGACCGAGUUCUAAUUGGAUUUUGGAGU-----UUGGAA-- (((((((((((((((.(((((.((((((((.((((.---...(((((.....))).))....)))---)))))))))..)).))).)))))))))))))...))...-----......-- ( -49.60) >DroEre_CAF1 4109 107 - 1 CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACC---ACGGGCAGUGAAACUGACCACAAGUG---CGGAGGGGGGAUGACCGAGUUCUAAUUGGAUUUUGGAGU-----UUGCAA-- (((((((((((((((.(((((.((((((((.((((.---...(((((.....))).))....)))---)))))))))..)).))).)))))))))))))...))...-----......-- ( -49.60) >DroYak_CAF1 3609 107 - 1 CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACC---ACGGGCAGUGAAACUGACCACAAGUG---CGGAGGGGGGAUGACCGACUUCUAAUUGGAUUUUGGAGU-----UUCGAA-- ((((((((((((((..(((((.((((((((.((((.---...(((((.....))).))....)))---)))))))))..)).)))..))))))))))))...))...-----......-- ( -46.80) >DroAna_CAF1 3042 120 - 1 CCGUCCAAUUAGAACAUGGCAACUCCCUCCAGCACGACCCCGGGCAGUGAGACUGACCCGAAGAGUGCCGGAGGGGGGCUGACCGAGUUCUAAUUGGAUUUCGCCGUCGAUUUGGGAGUA ..(((((((((((((.(((((.((((((((.((((.....(((((((.....))).))))....)))).))))))))..)).))).)))))))))))))(((.(((......)))))).. ( -60.60) >consensus CCGUCCAAUUAGAAUAUGGCGACUCCCUCCAGCACC___ACGGGCAGUGAGACUGACCACAAGUG___CGGAGGGGGGAUGACCGAGUUCUAAUUGGAUUUUGGAGU_____UUGGAA__ ..(((((((((((((.(((((.((((((((..(((......((.(((.....))).))....)))....))))))))..)).))).)))))))))))))..................... (-41.06 = -41.15 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:38 2006