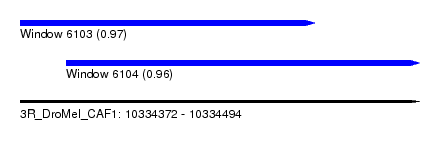
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,334,372 – 10,334,494 |
| Length | 122 |
| Max. P | 0.967731 |

| Location | 10,334,372 – 10,334,462 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 91.14 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -20.46 |
| Energy contribution | -19.43 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
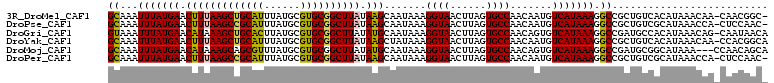
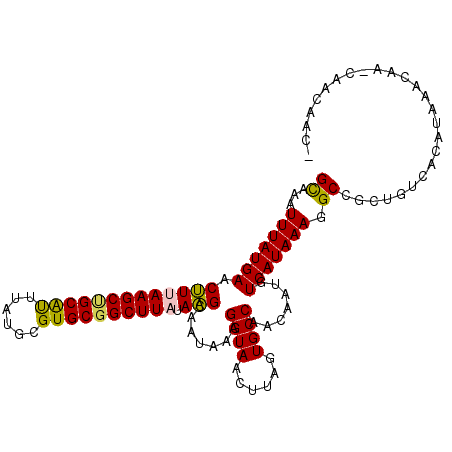
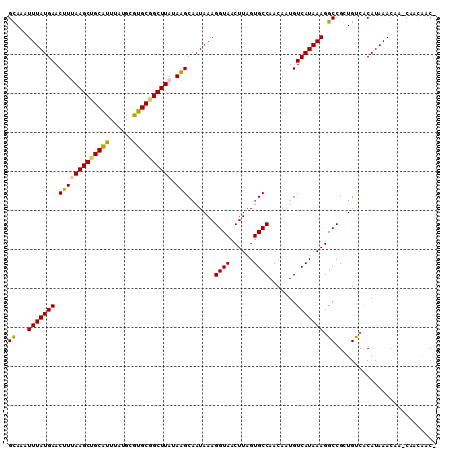
>3R_DroMel_CAF1 10334372 90 + 27905053 CCGAGUGAAUAUUGGCAAAUUUAUGAACUUUAAGCUGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUC ........((((((.....(((((...(((((((((((((......)))))))))).)))..)))))((((......))))..)))))). ( -23.00) >DroVir_CAF1 214436 85 + 1 -----CGAGUAUUGGCAAAUUUAUGAACAUAAAGCUGCAUUUAUGCGUGCGGCUUAUAUGCAAUAAAGGUAACUUAGUGCCAACAGUGUC -----......((((((..(((((...(((((((((((((......))))))))).))))..)))))..........))))))....... ( -22.60) >DroPse_CAF1 208216 88 + 1 --GGCCGAAUAUUGGCAAAUUUAUGAACUUUAAGCCGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUC --.(((((...)))))...(((((...(((((((((((((......)))))))))).)))..)))))((((......))))......... ( -26.50) >DroGri_CAF1 209349 85 + 1 -----CGAGUAUUGGUAAAUUUAUGAACAUAAAGCUGCACUUAUGCGUGCGGCUUAUAUGCAAUAAAGGUAACUUAGUGCCAACAGUGUC -----....(((((.....(((((...(((((((((((((......))))))))).))))..)))))((((......))))..))))).. ( -24.30) >DroMoj_CAF1 221177 85 + 1 -----CGAGUAUUGGCAAAUUUAUGAACAUAAAGCAGCGUUUAUGCGUGCGGCUUAUAUGCAAUAAAGGUAACUUAGUGCCAACAGUGUC -----......((((((..(((((...((((((((.((((....))))...)))).))))..)))))..........))))))....... ( -18.90) >DroPer_CAF1 212111 88 + 1 --GGCCGAAUAUUGGCAAAUUUAUGAACUUUAAGCCGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUC --.(((((...)))))...(((((...(((((((((((((......)))))))))).)))..)))))((((......))))......... ( -26.50) >consensus _____CGAAUAUUGGCAAAUUUAUGAACAUAAAGCUGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUC .........(((((.....(((((...(((..((((((((......))))))))...)))..)))))((((......))))..))))).. (-20.46 = -19.43 + -1.03)

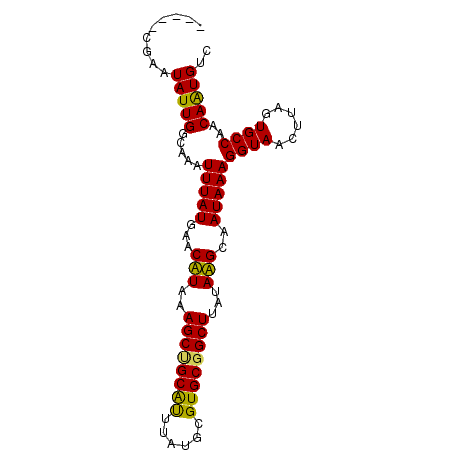
| Location | 10,334,386 – 10,334,494 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.00 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10334386 108 + 27905053 GCAAAUUUAUGAACUUUAAGCUGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUCAUAAAGGCCGCUGUCACAUAAACAA-CAACGGC- .....(((((((.(((((((((((((......)))))))))).))).......((((......)))).......))))))).((((.(((..........)-)).))))- ( -28.60) >DroPse_CAF1 208228 108 + 1 GCAAAUUUAUGAACUUUAAGCCGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUCAUAAAGGCCGCUGUCGCAUAAACCA-CUCCAAC- ((...(((((((.(((((((((((((......)))))))))).))).......((((......)))).......))))))).)).((....))........-.......- ( -28.40) >DroGri_CAF1 209358 109 + 1 GUAAAUUUAUGAACAUAAAGCUGCACUUAUGCGUGCGGCUUAUAUGCAAUAAAGGUAACUUAGUGCCAACAGUGUCAUAAAGGCCGAUGCCACAUAAACAG-CAAUAACA ((...(((((((.(((((((((((((......))))))))).)))).......((((......)))).......)))))))(((....))).........)-)....... ( -28.80) >DroYak_CAF1 201815 109 + 1 GCAAAUUUAUGAACUUUAAGCUGCAUUUAUGCGUGCGGCUUAUAAGCUAUAAAGGUAACUUAGUGCCAACAAUGUCAUAAAGGCCGCUGUCACAUAAACAA-CCACGGCA ((...(((((((.(((((((((((((......)))))))))).))).......((((......)))).......))))))).)).(((((...........-..))))). ( -29.42) >DroMoj_CAF1 221186 107 + 1 GCAAAUUUAUGAACAUAAAGCAGCGUUUAUGCGUGCGGCUUAUAUGCAAUAAAGGUAACUUAGUGCCAACAGUGUCAUAAAGGCCGAUGCGGCAUAAA---CCAACAGCA ((...(((((....)))))...))(((((((((((((((((.((((((.....((((......)))).....)).)))).)))))).))).)))))))---)........ ( -31.70) >DroPer_CAF1 212123 108 + 1 GCAAAUUUAUGAACUUUAAGCCGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUCAUAAAGGCCGCUGUCGCAUAAACCA-CUCCAAC- ((...(((((((.(((((((((((((......)))))))))).))).......((((......)))).......))))))).)).((....))........-.......- ( -28.40) >consensus GCAAAUUUAUGAACUUUAAGCUGCAUUUAUGCGUGCGGCUUAUAAGCAAUAAAGGUAACUUAGUGCCAACAAUGUCAUAAAGGCCGCUGUCACAUAAACAA_CAACAAC_ ((...(((((((.(((((((((((((......)))))))))).))).......((((......)))).......))))))).)).......................... (-24.52 = -24.00 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:35 2006