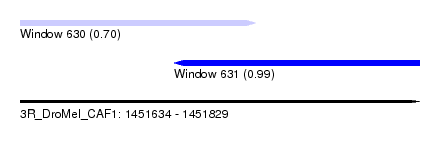
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,451,634 – 1,451,829 |
| Length | 195 |
| Max. P | 0.985967 |

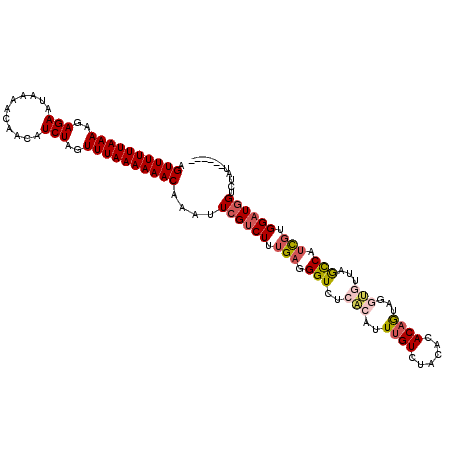
| Location | 1,451,634 – 1,451,749 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.90 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1451634 115 + 27905053 AGUUUUUUAAAAGAGAAUAAAACAACAUCUAGUUUAAAAAACAAAUUCGUCUUUGAGGGUCUCCUAUUUGUUUGCACACAGUAGUGAUUAGCCAUCGUGGAUGGACUUUUUCUUA .((((((((((..(((...........)))..))))))))))...(((((((.(((.(((.((((((((((....))).))))).))...))).))).))))))).......... ( -25.70) >DroSec_CAF1 24232 115 + 1 AGUUUUUUAAAAGAGAAUAAAACAACAUCUAGUUUAAAAAACAAAUUCGUCUUUGAGGGUCUCACAUUUGUCUACACACAAAAGGUGUUAGCCAUCGUGGAUGGUCUAUUGUUUA .((((((((((..(((...........)))..))))))))))....((((((.(((.(((..(((.(((((......)))))..)))...))).))).))))))........... ( -25.70) >DroSim_CAF1 22547 108 + 1 AGUUUUUUAAAAGAGAAUAAAACAACAUCUAGUUUAAAAAACAAAUUCGUCUUUGAGGGUCUCACAUUUGUCUGCACACAGUAGGUGUUAGCCAUCGUGGAUGGUCUA------- .((((((((((..(((...........)))..))))))))))....((((((.(((.((.((.(((((((.(((....)))))))))).)))).))).))))))....------- ( -29.00) >DroEre_CAF1 25750 103 + 1 AGUUUUUUAAAAGAGAAUAAAACAACAUCUAGUUUAAAAAACAAAUUCGUCUUUGAGGGUCUCAUAUUUGUUUACAAACUG------CUGAUCAGUUUGGCUUGUAUAU------ .((((((((((..(((...........)))..))))))))))...((((....))))......((((..((...(((((((------.....)))))))))..))))..------ ( -17.90) >consensus AGUUUUUUAAAAGAGAAUAAAACAACAUCUAGUUUAAAAAACAAAUUCGUCUUUGAGGGUCUCACAUUUGUCUACACACAGUAGGUGUUAGCCAUCGUGGAUGGUCUAU______ .((((((((((..(((...........)))..))))))))))....((((((.(((.(((..(((..((((......))))...)))...))).))).))))))........... (-16.40 = -17.90 + 1.50)

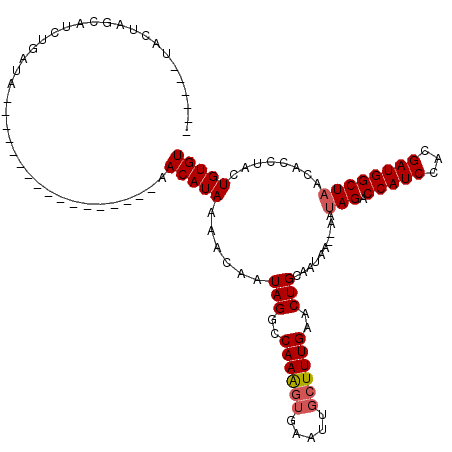
| Location | 1,451,709 – 1,451,829 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -11.35 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1451709 120 - 27905053 AGGGUAUACUAGAAUCUGAUAAGCAUAUCAAGAUUCAAAACAUAAAAGAAUAGGACAAGGUGAAGUCAUUUGAACUGCAAUAAGAAAAAGUCCAUCCACGAUGGCUAAUCACUACUGUGU .((((......((((((((((....)))).))))))................((((.(((((....)))))...((......)).....))))))))............(((....))). ( -22.40) >DroSec_CAF1 24307 97 - 1 ------UACUAGCAUCUGAUA-----------------AACAUAAAACAAUAGUCCAAAGUGAAUUGCUUUGAACUGCAAUAAACAAUAGACCAUCCACGAUGGCUAACACCUUUUGUGU ------...............-----------------.((((((((...((((.((((((.....)))))).))))..........(((.(((((...)))))))).....)))))))) ( -19.60) >DroSim_CAF1 22622 90 - 1 ------UACUAGCAUAUGAUA-----------------AACAUAAAACAAUAGGCCAAAGUAAAUUGCUUUGGACUGCAA-------UAGACCAUCCACGAUGGCUAACACCUACUGUGU ------........((((...-----------------..))))..(((.(((((((((((.....))))))).......-------(((.(((((...))))))))...)))).))).. ( -21.50) >consensus ______UACUAGCAUCUGAUA_________________AACAUAAAACAAUAGGCCAAAGUGAAUUGCUUUGAACUGCAAUAA__AAUAGACCAUCCACGAUGGCUAACACCUACUGUGU .......................................(((((......(((..((((((.....))))))..)))..........(((.(((((...))))))))........))))) (-11.35 = -12.13 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:50 2006