
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,264,240 – 10,264,410 |
| Length | 170 |
| Max. P | 0.997087 |

| Location | 10,264,240 – 10,264,332 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
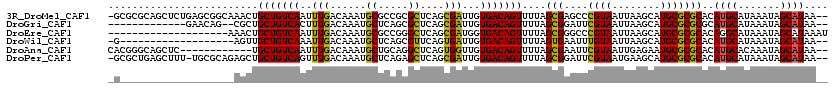
>3R_DroMel_CAF1 10264240 92 + 27905053 -------CUCCUGCUCGCUAAUGUCUUGGCCAAGAUUAAGCGAAAAAGUGCGCAAUCUUUGCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACA -------.....(((((((...(((((....)))))..)))))....(((((((.....))))))).))....(((((....)))))(((.....))). ( -30.90) >DroPse_CAF1 131548 81 + 1 U----UCUU-CUGGG------------CGUCAAGAUUAAGCGCAAAAGUGCGCAAUCUGUGCGCUGAGCUUU-UGCGCAGAGCUGCUGUCAGUUUGACA .----....-.....------------.(((........(((((((((((((((.....)))))...)))))-))))).((((((....))))))))). ( -30.50) >DroSec_CAF1 121765 95 + 1 U----UCCUCCUGCUCGCCAAAGUCUUGGCCAAGAUUAAGCUAAAAAGUCCGCAAUCUUUGCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACA .----.....(((((((((((....)))))........((((.....((.((((.....)))).))))))..)))))).........(((.....))). ( -24.40) >DroSim_CAF1 120963 95 + 1 U----UCCUCCUGCUCGCCAAAGUCUUGGCCAAGAUUAAGCGAAAAAGUGCGCAAUCUUUGCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACA .----.......((((((...((((((....))))))..))))....(((((((.....))))))).))....(((((....)))))(((.....))). ( -29.40) >DroYak_CAF1 129233 99 + 1 UCCUUGCCUUCUCCACACCAAAGUCUUGGCCAAGAUUAAGCGAAAAAGUGCGCAAUCUUUGAGCGCAACUCUGAGCGGCAAACUGCUGUCAAUUUGACA ...(((((.............((((((....))))))..((.......(((((.........))))).......)))))))......(((.....))). ( -23.34) >DroPer_CAF1 134172 81 + 1 U----UCUU-CUGGG------------CGUCAAGAUUAAGCGCAAAAGUGCGCAAUCUGUGCGCUGAGCUUU-UGCGCAGAGCUGCUGUCAGUUUGACA .----....-.....------------.(((........(((((((((((((((.....)))))...)))))-))))).((((((....))))))))). ( -30.50) >consensus U____UCCUCCUGCUCGCCAAAGUCUUGGCCAAGAUUAAGCGAAAAAGUGCGCAAUCUUUGCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACA ......................................(((........(((((.....)))))((.((.....)).)).....)))(((.....))). (-14.65 = -14.27 + -0.39)



| Location | 10,264,240 – 10,264,332 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -16.85 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10264240 92 - 27905053 UGUCAAAUUGACAGCAGUUUGCCGCUCAGAGCUGCGCGCAAAGAUUGCGCACUUUUUCGCUUAAUCUUGGCCAAGACAUUAGCGAGCAGGAG------- ((((.....))))(((((((........)))))))(((((.....))))).(((.((((((...((((....))))....)))))).)))..------- ( -31.40) >DroPse_CAF1 131548 81 - 1 UGUCAAACUGACAGCAGCUCUGCGCA-AAAGCUCAGCGCACAGAUUGCGCACUUUUGCGCUUAAUCUUGACG------------CCCAG-AAGA----A .(((((.(((....)))....(((((-((((....(((((.....))))).)))))))))......))))).------------.....-....----. ( -25.80) >DroSec_CAF1 121765 95 - 1 UGUCAAAUUGACAGCAGUUUGCCGCUCAGAGCUGCGCGCAAAGAUUGCGGACUUUUUAGCUUAAUCUUGGCCAAGACUUUGGCGAGCAGGAGGA----A ((((.....)))).(..((((((((.(((((((..(.((.(((((((.((.((....))))))))))).))).)).)))))))).)))))..).----. ( -28.40) >DroSim_CAF1 120963 95 - 1 UGUCAAAUUGACAGCAGUUUGCCGCUCAGAGCUGCGCGCAAAGAUUGCGCACUUUUUCGCUUAAUCUUGGCCAAGACUUUGGCGAGCAGGAGGA----A ((((.....))))(((((((........)))))))(((((.....))))).(((.((((((...((((....))))....)))))).)))....----. ( -31.40) >DroYak_CAF1 129233 99 - 1 UGUCAAAUUGACAGCAGUUUGCCGCUCAGAGUUGCGCUCAAAGAUUGCGCACUUUUUCGCUUAAUCUUGGCCAAGACUUUGGUGUGGAGAAGGCAAGGA ((((.....))))((..(((.((((((((((((..(((..(((((((.((........)).))))))))))...)))))))).)))))))..))..... ( -32.20) >DroPer_CAF1 134172 81 - 1 UGUCAAACUGACAGCAGCUCUGCGCA-AAAGCUCAGCGCACAGAUUGCGCACUUUUGCGCUUAAUCUUGACG------------CCCAG-AAGA----A .(((((.(((....)))....(((((-((((....(((((.....))))).)))))))))......))))).------------.....-....----. ( -25.80) >consensus UGUCAAAUUGACAGCAGUUUGCCGCUCAGAGCUGCGCGCAAAGAUUGCGCACUUUUUCGCUUAAUCUUGGCCAAGACUUUGGCGAGCAGGAGGA____A .(((((..(...(((.....((.((.....)).))(((((.....)))))........)))..)..)))))............................ (-16.85 = -15.93 + -0.91)



| Location | 10,264,255 – 10,264,372 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -17.67 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10264255 117 + 27905053 GUCUUGGCCAAGAUUAAGCGAAAAA-GUGCGCAAUCUUUGCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACAAAUGCGCCGCGCUCAGCGAUUGUGACAGUUUUAGCGAGCC (((((....)))))...((..((((-.(((((((((.(((.((((.((....(((((....)))))(((.....)))......)).)))).))).))))))).)).)))).))..... ( -39.00) >DroVir_CAF1 138950 102 + 1 GCGCCUGUCAAGAUUAAGCUCAAGAGCUGCGCAAUCGCU------------GAGCGG----CUGCUGUCAAUUUGACAAAUGCUCAGCUCUCAGCGAUUGUGACAGUUUUAGCGGAUU .................(((..(((((((((((((((((------------(((.((----(((.((((.....))))......)))))))))))))))))).))))))))))..... ( -42.20) >DroPse_CAF1 131558 111 + 1 -----CGUCAAGAUUAAGCGCAAAA-GUGCGCAAUCUGUGCGCUGAGCUUU-UGCGCAGAGCUGCUGUCAGUUUGACAAAUGCUCAGAGCUCAGCGAUUGUGACAGUUUUAGCGGAUU -----.(((((((((..(((((...-.))))))))))...(((((((((((-.(((((....)))((((.....))))...))..)))))))))))....)))).............. ( -41.40) >DroWil_CAF1 135405 98 + 1 CUCUUUCACAAGAUUAAGCGCAAAA-GUGCGCAAUCUUAG-------------------AGUUGCUGUCAAAUUGACAAAUGCUCAGCGUUCAGUGAUUGUGACAGUUUUAGUGAAUU ....(((((((((((..(((((...-.)))))))))))..-------------------....(((((((.(((((...((((...))))))))).....)))))))....))))).. ( -28.60) >DroMoj_CAF1 135271 102 + 1 GCUCUCGUCAAGAUUAAGCGCAAAAGCUGCGCAAUCGCU------------GAGCGG----CUGCUGUCAAUUUGACAAAUGCUCAGCGCUCAGCGAUUGUGACAGUUUUAGCGGAUU (((.((.....))...)))((.(((((((((((((((((------------(((((.----(((.((((.....))))......)))))))))))))))))).))))))).))..... ( -47.10) >DroPer_CAF1 134182 111 + 1 -----CGUCAAGAUUAAGCGCAAAA-GUGCGCAAUCUGUGCGCUGAGCUUU-UGCGCAGAGCUGCUGUCAGUUUGACAAAUGCUCAGAGCUCAGCGAUUGUGACAGUUUUAGCGGAUU -----.(((((((((..(((((...-.))))))))))...(((((((((((-.(((((....)))((((.....))))...))..)))))))))))....)))).............. ( -41.40) >consensus G___UCGUCAAGAUUAAGCGCAAAA_GUGCGCAAUCUCUG____________AGCGG__AGCUGCUGUCAAUUUGACAAAUGCUCAGCGCUCAGCGAUUGUGACAGUUUUAGCGGAUU ...........((((..(((((.....))))))))).........................((((((......(.((((.((((........)))).)))).)......))))))... (-17.67 = -17.83 + 0.17)


| Location | 10,264,255 – 10,264,372 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -14.08 |
| Energy contribution | -15.13 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10264255 117 - 27905053 GGCUCGCUAAAACUGUCACAAUCGCUGAGCGCGGCGCAUUUGUCAAAUUGACAGCAGUUUGCCGCUCAGAGCUGCGCGCAAAGAUUGCGCAC-UUUUUCGCUUAAUCUUGGCCAAGAC ((((..................(((.....)))((((...((((.....))))(((((((........))))))))))).(((((((.((..-......)).)))))))))))..... ( -36.20) >DroVir_CAF1 138950 102 - 1 AAUCCGCUAAAACUGUCACAAUCGCUGAGAGCUGAGCAUUUGUCAAAUUGACAGCAG----CCGCUC------------AGCGAUUGCGCAGCUCUUGAGCUUAAUCUUGACAGGCGC ....(((.....(((((((((((((((((.((((.....(((((.....))))))))----)..)))------------))))))))...((((....))))......))))))))). ( -39.00) >DroPse_CAF1 131558 111 - 1 AAUCCGCUAAAACUGUCACAAUCGCUGAGCUCUGAGCAUUUGUCAAACUGACAGCAGCUCUGCGCA-AAAGCUCAGCGCACAGAUUGCGCAC-UUUUGCGCUUAAUCUUGACG----- ..............((((....((((((((((.((((..(((((.....)))))..)))).)....-..)))))))))...((((((((((.-...)))))..))))))))).----- ( -37.30) >DroWil_CAF1 135405 98 - 1 AAUUCACUAAAACUGUCACAAUCACUGAACGCUGAGCAUUUGUCAAUUUGACAGCAACU-------------------CUAAGAUUGCGCAC-UUUUGCGCUUAAUCUUGUGAAAGAG ............((.((((..............(((...(((((.....)))))...))-------------------).(((((((((((.-...)))))..)))))))))).)).. ( -22.70) >DroMoj_CAF1 135271 102 - 1 AAUCCGCUAAAACUGUCACAAUCGCUGAGCGCUGAGCAUUUGUCAAAUUGACAGCAG----CCGCUC------------AGCGAUUGCGCAGCUUUUGCGCUUAAUCUUGACGAGAGC .....(((.....(((((.(((((((((((((((.....(((((.....))))))))----.)))))------------)))))))((((((...)))))).......)))))..))) ( -40.40) >DroPer_CAF1 134182 111 - 1 AAUCCGCUAAAACUGUCACAAUCGCUGAGCUCUGAGCAUUUGUCAAACUGACAGCAGCUCUGCGCA-AAAGCUCAGCGCACAGAUUGCGCAC-UUUUGCGCUUAAUCUUGACG----- ..............((((....((((((((((.((((..(((((.....)))))..)))).)....-..)))))))))...((((((((((.-...)))))..))))))))).----- ( -37.30) >consensus AAUCCGCUAAAACUGUCACAAUCGCUGAGCGCUGAGCAUUUGUCAAAUUGACAGCAGCU__CCGCU____________CAAAGAUUGCGCAC_UUUUGCGCUUAAUCUUGACGA___C ..............((((.(((((((.(....).)))..(((((.....)))))............................))))(((((.....))))).......))))...... (-14.08 = -15.13 + 1.06)


| Location | 10,264,293 – 10,264,410 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -16.98 |
| Energy contribution | -16.37 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10264293 117 + 27905053 -GCGCGCAGCUCUGAGCGGCAAACUGCUGUCAAUUUGACAAAUGCGCCGCGCUCAGCGAUUGUGACAGUUUUAGCGAGCCCGUAAUUAAGCAUGCGCGCACAUGCAUAAAUAGCAUAA-- -((((((((((.((((((((.....((((((.....))))...)))))))((((.(((((((...)))))...))))))......)))))).)))))))..((((.......))))..-- ( -40.70) >DroGri_CAF1 127469 103 + 1 -------------GAACAG--CGCUGCUGUCACUUUGACAAAUGCUCAGCGCUCAGCGAUUGUGACAGUUUUAGCGGAUUCGUAAUUAAGCAUGCGCGCGCAUGCAUAAAUAGCAUAA-- -------------.....(--(.((((((..(((.(.((((.((((........)))).)))).).)))..))))))............((((((....)))))).......))....-- ( -30.50) >DroEre_CAF1 122443 100 + 1 --------------------AAACUGCUGUCAAUUUGACAAAUGCGCCGGGCUCAGCGAUGGUGACAGUUUUAGCGGGCCCGUAAUUAAGCAUGCGCGCACAGGCAUAAAUAGCAUAAAU --------------------...(((.((((.....))))..(((((.((((((.((..((....))......))))))))(((........)))))))))))((.......))...... ( -31.30) >DroWil_CAF1 135443 98 + 1 -G-------------------AGUUGCUGUCAAAUUGACAAAUGCUCAGCGUUCAGUGAUUGUGACAGUUUUAGUGAAUUUGUAAUUAAGCAUGCGCGCACAUGCAUAAAUAGCAUAA-- -(-------------------(((...((((.....))))...)))).((((.((.((((((..(..((((....)))))..))))))....)).))))..((((.......))))..-- ( -23.10) >DroAna_CAF1 109814 106 + 1 CACGGGCAGCUC------------UGCUGUCAAUUUGACAAAUGCUGCAGGCUCAGUGGUUGUGACAGUUUUAGCGAAUUCGUAAUUGAGAAUGCGCGCACAUGCACAAAUAGCAUAA-- .....(((((..------------...((((.....))))...))))).......((..(((((.((......(((.((((........)))).))).....)))))))...))....-- ( -28.30) >DroPer_CAF1 134215 116 + 1 -GCGCUGAGCUUU-UGCGCAGAGCUGCUGUCAGUUUGACAAAUGCUCAGAGCUCAGCGAUUGUGACAGUUUUAGCGGAUUCGUAAUGAAGCAUGCGCGCACAUGCAUAAAUAGCAUAA-- -((((((((((((-.(((((....)))((((.....))))...))..))))))))))....(((.(((((((((((....)))..)))))).)))))))..((((.......))))..-- ( -39.20) >consensus _G___________________AGCUGCUGUCAAUUUGACAAAUGCUCAGCGCUCAGCGAUUGUGACAGUUUUAGCGAAUUCGUAAUUAAGCAUGCGCGCACAUGCAUAAAUAGCAUAA__ .........................(((((((..(((......((.....))....)))...)))))))....(((....((((........)))))))..((((.......)))).... (-16.98 = -16.37 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:06 2006