| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,250,999 – 10,251,091 |
| Length | 92 |
| Max. P | 0.880568 |

| Location | 10,250,999 – 10,251,091 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -20.31 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10250999 92 + 27905053 GUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUUGCCGGCUAAUAAUUUUAAUUUAGCCA----AAA----------------------AAUUAGUUGUAUUCGUUGGU ...(((((((-((((((((.....)))))).....)))...))))))((((((..........)))))).----...----------------------.................... ( -23.30) >DroVir_CAF1 125999 90 + 1 GUGGGCAGCA-CUGUGACACAUUUUGUCAUCAUUAGCCACAGUUACCGGCUAAUAAUUUCAAUUUAGCCA----AAA----------------------AAUUAGCUGCAUUC--AGCU ....((((((-(((((.....................))))))....((((((..........)))))).----...----------------------.....)))))....--.... ( -22.40) >DroGri_CAF1 114311 112 + 1 GUGGGCAGCA-GUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCCA----AAAAAAAAAAAAAAGAAGAAAAUAAAAUUAGCCGCAUUG--AGCU (((((((((.-((((((((.....)))))........))).))))))((((((..........)))))).----................................)))....--.... ( -25.20) >DroWil_CAF1 122268 94 + 1 GUGGGCAGCGGCUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUGGCCAACAAAAA----------------------AAUUAGUUGCAUAUAUU--- (((((((((((((((((((.....))))))....))))...))))))((((((..........))))))........----------------------.........))).....--- ( -27.30) >DroMoj_CAF1 119515 90 + 1 GUGGGCAGCA-CUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCCA----AAA----------------------AAUUAGCUGCAUUG--AGCU ....((((((-(((((.....................))))))....((((((..........)))))).----...----------------------.....)))))....--.... ( -22.40) >DroAna_CAF1 98179 92 + 1 GUGGGCAGCG-CUGUGACACAUUUUGUCAUCAUUAGGCACAGUCGCCGGCUAAUAAUUUCAAUUUGGCCA----AAA----------------------AAUUAGUUGCAUUUCGGAGU ....(((((.-..((((((.....)))))).....(((......)))((((((..........)))))).----...----------------------.....))))).......... ( -23.80) >consensus GUGGGCAGCA_CUGUGACACAUUUUGUCAUCAUUAGCCACAGUUGCCGGCUAAUAAUUUCAAUUUAGCCA____AAA______________________AAUUAGCUGCAUUC__AGCU ....(((((....((((((.....)))))).................((((((..........))))))...................................))))).......... (-20.31 = -19.87 + -0.44)

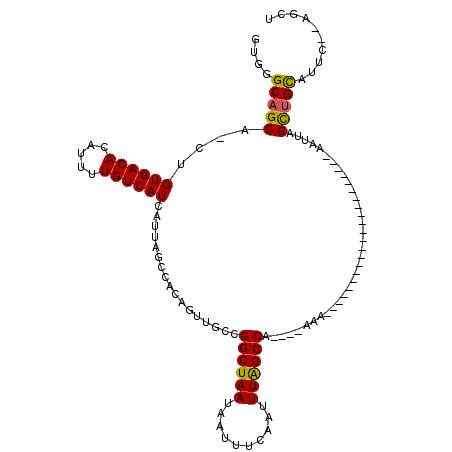

| Location | 10,250,999 – 10,251,091 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -16.52 |
| Energy contribution | -17.55 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10250999 92 - 27905053 ACCAACGAAUACAACUAAUU----------------------UUU----UGGCUAAAUUAAAAUUAUUAGCCGGCAACUGUGCCUAAUGAUGACAAAAUGUGUCACAG-CGCUGCCCAC ....................----------------------...----.((((((..........))))))((((...((((....((.(((((.....))))))))-)))))))... ( -19.90) >DroVir_CAF1 125999 90 - 1 AGCU--GAAUGCAGCUAAUU----------------------UUU----UGGCUAAAUUGAAAUUAUUAGCCGGUAACUGUGGCUAAUGAUGACAAAAUGUGUCACAG-UGCUGCCCAC ((((--(....)))))....----------------------...----.(((...((((..(((((((((((.......)))))))))))((((.....)))).)))-)...)))... ( -28.20) >DroGri_CAF1 114311 112 - 1 AGCU--CAAUGCGGCUAAUUUUAUUUUCUUCUUUUUUUUUUUUUU----UGGCUAAAUUGAAAUUAUUAGCCGGCAACUGUGGCUAAUGAUGACAAAAUGUGUCACAC-UGCUGCCCAC ((((--((((..((((((..........................)----)))))..))))).(((((((((((.(....))))))))))))((((.....))))....-.)))...... ( -25.57) >DroWil_CAF1 122268 94 - 1 ---AAUAUAUGCAACUAAUU----------------------UUUUUGUUGGCCAAAUUGAAAUUAUUAGCCGGCAACUGUGGCUAAUGAUGACAAAAUGUGUCACAGCCGCUGCCCAC ---.......(((.......----------------------...((((((((..(((.......))).))))))))..((((((...(.(((((.....))))))))))))))).... ( -22.50) >DroMoj_CAF1 119515 90 - 1 AGCU--CAAUGCAGCUAAUU----------------------UUU----UGGCUAAAUUGAAAUUAUUAGCCGGCAACUGUGGCUAAUGAUGACAAAAUGUGUCACAG-UGCUGCCCAC ((((--((((..((((((..----------------------..)----)))))..))))).(((((((((((.(....))))))))))))((((.....))))....-.)))...... ( -27.50) >DroAna_CAF1 98179 92 - 1 ACUCCGAAAUGCAACUAAUU----------------------UUU----UGGCCAAAUUGAAAUUAUUAGCCGGCGACUGUGCCUAAUGAUGACAAAAUGUGUCACAG-CGCUGCCCAC ...((((((...........----------------------)))----)))...................(((((.((((..(....)..((((.....))))))))-)))))..... ( -15.60) >consensus AGCU__AAAUGCAACUAAUU______________________UUU____UGGCUAAAUUGAAAUUAUUAGCCGGCAACUGUGGCUAAUGAUGACAAAAUGUGUCACAG_CGCUGCCCAC ..........((......................................((((((..........))))))((((.(((((((.................))))))).)))))).... (-16.52 = -17.55 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:59 2006