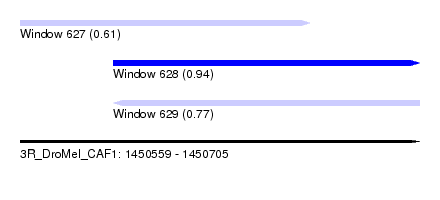
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,450,559 – 1,450,705 |
| Length | 146 |
| Max. P | 0.939882 |

| Location | 1,450,559 – 1,450,665 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.31 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -24.16 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1450559 106 + 27905053 ACAGUGCUCGAGU-UGUU-UUCGUAUUAAACUUGAAUUUCCCUUUCAAUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGUCGCCUGGCCUCCGUGGUG .(((.((((((((-(...-.........)))))))..................(((((..((....((((((....)))))).)).))))))))))(((.....))). ( -30.60) >DroSec_CAF1 23143 106 + 1 ACUGUGCUCGAGU-UGUU-UUUGUAGUUAACUUGAAUUUCCCUUUAAAUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGCCGCCUGGCCUCCGUGGUG ((..((.((((((-((..-........)))))))).....((...........(((((((......)))))))((((((........))))))..))....))..)). ( -31.90) >DroSim_CAF1 21463 106 + 1 ACAGUGCUCGAGU-UGUU-UUUGUAGUUAACUUGAAUUUCCCUUUAAAUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGCCGCCUGGCCUCCGUGGUG .(((...((((((-((..-........))))))))..................(((((((......)))))))((((((........)))))))))(((.....))). ( -33.00) >DroEre_CAF1 24675 104 + 1 ACAGUGCUCGAAU-UGUU-UUCUAAUUUAACUUAAAUUUU--UUUCCACUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGCCGCCUGGCCUCUGUGGUG ((((.(((.(((.-((..-....((((((...))))))..--....)).))).(((((((......)))))))((((((........))))))..)))..)))).... ( -29.60) >DroYak_CAF1 23196 107 + 1 ACCGAAUCCGAAU-UGUUUUUCUUAUUUAACUUAAAUUUCCCUUUUAACUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGCCGCCUGGCCUCCGUGGUG (((((((..(((.-.....)))..))))............((...........(((((((......)))))))((((((........))))))..)).......))). ( -26.40) >DroAna_CAF1 24431 98 + 1 ACAA-ACUGGAUUCUGUU-UUAAUAAGAGACU--------CUUUUUUUUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGUUGGGCCGUCUGGCCUCCGUGGUC ..((-(..(((.(((.((-.....)).))).)--------))..)))......(((((((......)))))))((((((........))))))..((((.....)))) ( -27.40) >consensus ACAGUGCUCGAGU_UGUU_UUCGUAUUUAACUUGAAUUUCCCUUUAAAUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGCCGCCUGGCCUCCGUGGUG .....................................................(((((((......)))))))((((((........))))))...(((.....))). (-24.16 = -23.88 + -0.28)

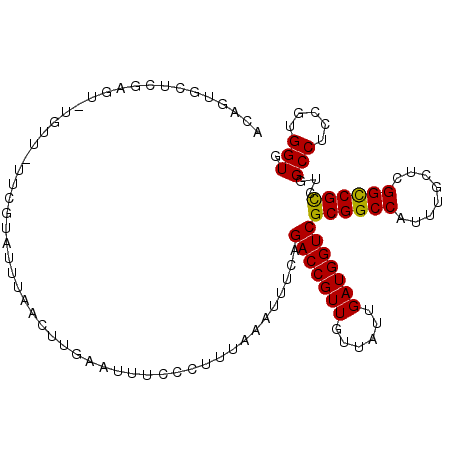
| Location | 1,450,593 – 1,450,705 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -45.52 |
| Consensus MFE | -37.90 |
| Energy contribution | -36.93 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1450593 112 + 27905053 UUUCCCUUU--------CAAUUUCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUCGGUCGCCUGGCCUCCGUGGUGGCCAAGUACCUGUUGCAGGGCGGCAAGGUGGCCGUGGUCC ....((...--------........(((((((......)))))))((((((((((.....(((((((((((.(....).))))..(((.....))).))))))).))))))))))))... ( -47.90) >DroVir_CAF1 23702 117 + 1 UU-GUUUUUCUA--UUUAAUUUUUAGACCGUUGUUAUUGAUGGACGCGGCCAUUUGCUUGGCCGCCUGGCCUCCGUGGUUGCCAAGUACCUGUUGCAGGGCGGCAAGGUGGCCGUAGUCC ..-.....((((--.........))))..............(((((((((((......)))))))..((((.((...((((((..(((.....)))..))))))..)).))))...)))) ( -44.70) >DroPse_CAF1 24941 104 + 1 --CCCC--------------UUGCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUGCUUGGCCGUCUGGCUUCCGUCGUCGCCAAGUACCUGUUGCAGGGCGGCAAGGUGGCCGUCGUUC --....--------------.....(((((((......)))))))(((((((......)))))))..((((.((...((((((..(((.....)))..))))))..)).))))....... ( -44.20) >DroGri_CAF1 23430 117 + 1 UUUGUCUUU--UUGCUU-GUCUGCAGACCGUUGUUAUUGAUGGUCGCGGCCAUUUACUGGGCCGCCUGGCCUCCGUUGUCGCCAAGUACCUGUUGCAGGGCGGCAAGGUAGCCGUGGUGC .........--..((..-..(..(.(((((((......)))))))((((((........))))))..(((..((.((((((((..(((.....)))..))))))))))..))))..).)) ( -48.90) >DroWil_CAF1 29012 109 + 1 UUUCA---U--------GUAUUGCAGACCGUUAUUAUUGAUGGACGCGGCCAUUUGCUGGGCCGUCUGGCCUCAGUUGUUGCCAAGUACCUGUUGCAGGGCGGCAAAGUGGCUGUGGUUC .....---.--------..........((((((....)))))).(((((((((((((((((((....)))).))))(((((((..(((.....)))..)))))))))))))))))).... ( -45.30) >DroMoj_CAF1 21823 110 + 1 UUUGUUUUU--A--------UUAUAGACCGUUGUUAUCGAUGGACGCGGCCAUUUGCUUGGCCGCCUGGCCUCCGUGGUAGCCAAGUACCUGUUGCAAGGCGGCAAGGUGGCUGUUGUGC .........--.--------.......((((((....))))))(((((((((..((((((((..(((((...))).))..))))))))((((((((...))))).))))))))).))).. ( -42.10) >consensus UUUCUCUUU___________UUGCAGACCGUUGUUAUUGAUGGACGCGGCCAUUUGCUUGGCCGCCUGGCCUCCGUGGUCGCCAAGUACCUGUUGCAGGGCGGCAAGGUGGCCGUGGUCC ...........................((((((....))))))..((((((((((.....((((((((((..(....)..)))..(((.....))).))))))).))))))))))..... (-37.90 = -36.93 + -0.97)
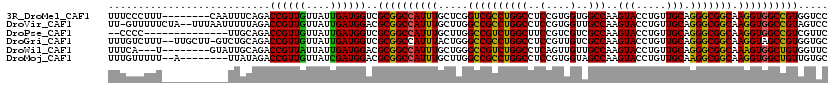
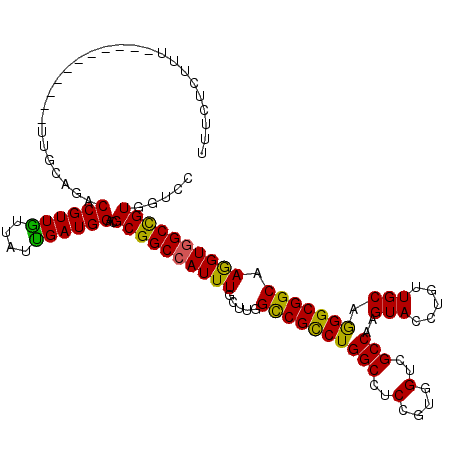
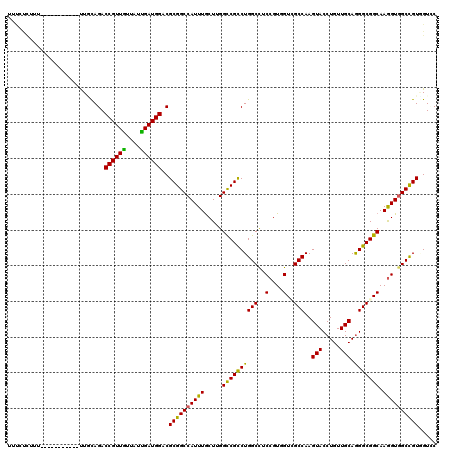
| Location | 1,450,593 – 1,450,705 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -24.61 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.773071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1450593 112 - 27905053 GGACCACGGCCACCUUGCCGCCCUGCAACAGGUACUUGGCCACCACGGAGGCCAGGCGACCGAGCAAAUGGCCGCGACCAUCAAUAACAACGGUCUGAAAUUG--------AAAGGGAAA ...((.((((((..((((((.((((...))))..(((((((.(....).)))))))....)).)))).)))))).((((............))))........--------...)).... ( -38.50) >DroVir_CAF1 23702 117 - 1 GGACUACGGCCACCUUGCCGCCCUGCAACAGGUACUUGGCAACCACGGAGGCCAGGCGGCCAAGCAAAUGGCCGCGUCCAUCAAUAACAACGGUCUAAAAAUUAAA--UAGAAAAAC-AA ((((...((((.((((((((.((((...))))....))))))....)).))))..(((((((......)))))))))))..............((((.........--)))).....-.. ( -40.90) >DroPse_CAF1 24941 104 - 1 GAACGACGGCCACCUUGCCGCCCUGCAACAGGUACUUGGCGACGACGGAAGCCAGACGGCCAAGCAAAUGGCCGCGACCAUCAAUAACAACGGUCUGCAA--------------GGGG-- ......((((......))))(((((((.........((((..(....)..))))(.((((((......)))))))((((............))))))).)--------------))).-- ( -35.40) >DroGri_CAF1 23430 117 - 1 GCACCACGGCUACCUUGCCGCCCUGCAACAGGUACUUGGCGACAACGGAGGCCAGGCGGCCCAGUAAAUGGCCGCGACCAUCAAUAACAACGGUCUGCAGAC-AAGCAA--AAAGACAAA ((....((((......))))..(((((...(((.((((....))).)...)))..((((((........))))))((((............)))))))))..-..))..--......... ( -38.50) >DroWil_CAF1 29012 109 - 1 GAACCACAGCCACUUUGCCGCCCUGCAACAGGUACUUGGCAACAACUGAGGCCAGACGGCCCAGCAAAUGGCCGCGUCCAUCAAUAAUAACGGUCUGCAAUAC--------A---UGAAA ......(((((...((((......))))..((.((.((((.....(((.((((....)))))))......)))).))))............)).)))......--------.---..... ( -28.00) >DroMoj_CAF1 21823 110 - 1 GCACAACAGCCACCUUGCCGCCUUGCAACAGGUACUUGGCUACCACGGAGGCCAGGCGGCCAAGCAAAUGGCCGCGUCCAUCGAUAACAACGGUCUAUAA--------U--AAAAACAAA .......(((((...((((...........))))..))))).......(((((..(((((((......)))))))(((....)))......)))))....--------.--......... ( -31.60) >consensus GAACCACGGCCACCUUGCCGCCCUGCAACAGGUACUUGGCAACCACGGAGGCCAGGCGGCCAAGCAAAUGGCCGCGACCAUCAAUAACAACGGUCUGCAA___________AAAGACAAA .......(((((((((((......)))..))))..(((((..(....)..)))))((((((........))))))................))))......................... (-24.61 = -25.17 + 0.56)

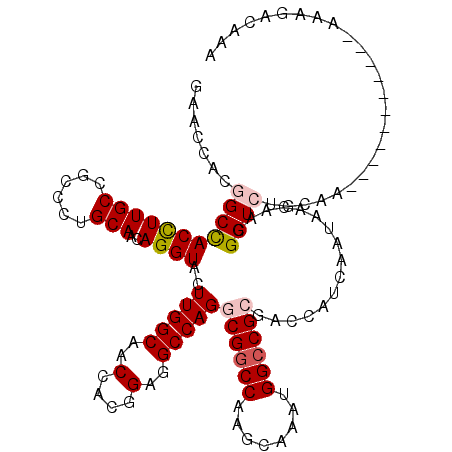
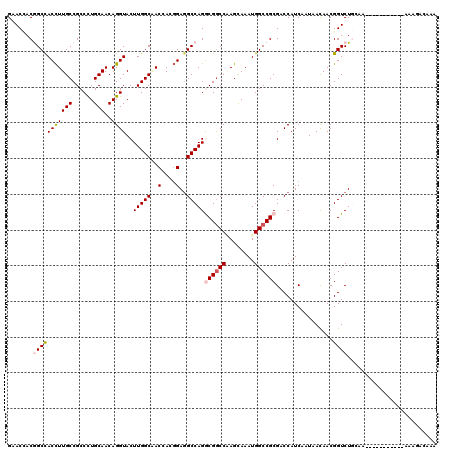
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:48 2006