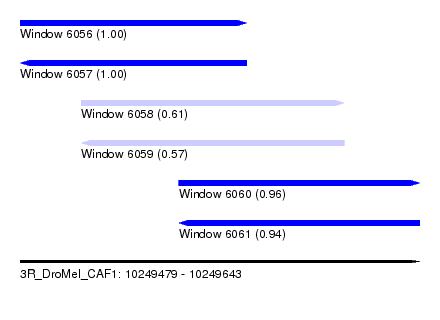
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,249,479 – 10,249,643 |
| Length | 164 |
| Max. P | 0.999898 |

| Location | 10,249,479 – 10,249,572 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -16.39 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249479 93 + 27905053 AACAUGCAACGCUGCAAUAUUGCAACAUUGCAACUCUGCGGCAUUGCAACUCUGCUGCAUUGCAAC--------UCUGCAACUGCAAUUGUAACGCGAUCA .((((((((.((((((...(((((....)))))...)))))).))))).......(((((((((..--------..))))).))))..))).......... ( -29.40) >DroSec_CAF1 107330 77 + 1 AACGUGCAACGCUGCAACAUUGCAACAUUGCAACUCUGCU----------------GCAUUGCAAC--------UCUGCAACUGCAAUUGUAACGCGAUCA ..((((..((...(((...(((((....)))))...)))(----------------((((((((..--------..))))).))))...))..)))).... ( -20.50) >DroSim_CAF1 105790 77 + 1 AACAUGCAACGCUGCAACAUUGCAACAUUGCAACUCUGCG----------------GCAUUGCAAC--------UCUGCAACUGCAAUUGUAACGCGAUCA .((((((((.((((((...(((((....)))))...))))----------------)).)))))..--------..(((....)))..))).......... ( -22.80) >DroYak_CAF1 113710 93 + 1 AACAUGCAACGCUGCAACAU--------UGCAACUCUGCAGCAUUGCAACUCUGCUGCAUUGCAACUCUGCGGCAUUGCAACUGCAAUUGUAACGCGAUCA ....(((...((((((...(--------(((((...(((((((.........)))))))))))))...))))))(((((....)))))......))).... ( -31.70) >consensus AACAUGCAACGCUGCAACAUUGCAACAUUGCAACUCUGCG________________GCAUUGCAAC________UCUGCAACUGCAAUUGUAACGCGAUCA .........((((((((..........((((((...(((((((.........)))))))))))))...........(((....))).)))))..))).... (-16.39 = -17.45 + 1.06)


| Location | 10,249,479 – 10,249,572 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.71 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249479 93 - 27905053 UGAUCGCGUUACAAUUGCAGUUGCAGA--------GUUGCAAUGCAGCAGAGUUGCAAUGCCGCAGAGUUGCAAUGUUGCAAUAUUGCAGCGUUGCAUGUU .....(((((.(((((((....)))..--------)))).)))))((((....((((((((.((((.((((((....)))))).)))).)))))))))))) ( -34.00) >DroSec_CAF1 107330 77 - 1 UGAUCGCGUUACAAUUGCAGUUGCAGA--------GUUGCAAUGC----------------AGCAGAGUUGCAAUGUUGCAAUGUUGCAGCGUUGCACGUU .....((((.....((((....)))).--------...(((((((----------------.((((.((((((....)))))).)))).))))))))))). ( -27.40) >DroSim_CAF1 105790 77 - 1 UGAUCGCGUUACAAUUGCAGUUGCAGA--------GUUGCAAUGC----------------CGCAGAGUUGCAAUGUUGCAAUGUUGCAGCGUUGCAUGUU ..........(((.((((....)))).--------..((((((((----------------.((((.((((((....)))))).)))).))))))))))). ( -26.60) >DroYak_CAF1 113710 93 - 1 UGAUCGCGUUACAAUUGCAGUUGCAAUGCCGCAGAGUUGCAAUGCAGCAGAGUUGCAAUGCUGCAGAGUUGCA--------AUGUUGCAGCGUUGCAUGUU .....(((.......)))...((((((((.((((.((((((((((((((.........)))))))...)))))--------)).)))).)))))))).... ( -37.50) >consensus UGAUCGCGUUACAAUUGCAGUUGCAGA________GUUGCAAUGC________________CGCAGAGUUGCAAUGUUGCAAUGUUGCAGCGUUGCAUGUU .....((((.......(((.((((((..........))))))))).................((((.((((((((((....)))))))))).)))))))). (-22.17 = -22.68 + 0.50)


| Location | 10,249,504 – 10,249,612 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -16.72 |
| Energy contribution | -16.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249504 108 + 27905053 CAUUGCAACUCUGCGGCAUUGCAACUCUGCUGCAUUGCAAC-UCUGCAA---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCU ....((((.......((...((.(((((((((.(((((.((-..(((..---..)))...))...)))))))))).)))).))..))((((.((....))))))...)))). ( -33.90) >DroVir_CAF1 124676 85 + 1 CA-GGCAACAG-G--------CAAC--------A-GGCAAC---UGCAA---CUGUAAC--UAACUCGAGCAGCAUGAGUAGCUGGCAAUAUUUGACAGCAGCUUAAUUGCU ..-((..((((-.--------...(--------(-(....)---))...---))))..)--)......(((((..(((((.((((.((.....)).)))).))))).))))) ( -23.30) >DroPse_CAF1 116214 89 + 1 CAUUGCAACUGU-----------------------UGUAACUGUUGCAACUGCUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUUUGACAGCGGCUUAAUUGCU ....(((...((-----------------------((...(((((((((..(((((.((((.....))))((((.......)))))))))..))).))))))..))))))). ( -26.00) >DroSec_CAF1 107355 92 + 1 CAUUGCAACUCUGCU----------------GCAUUGCAAC-UCUGCAA---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCU ....((.((((((((----------------(.(((((.((-..(((..---..)))...))...)))))))))).)))).)).(((((((.((....))))).....)))) ( -28.00) >DroSim_CAF1 105815 92 + 1 CAUUGCAACUCUGCG----------------GCAUUGCAAC-UCUGCAA---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCU ..((((((...((((----------------(..(((((..-..)))))---))))).)))))).(((((((((.......))))..((((.((....))))))..))))). ( -26.30) >DroEre_CAF1 107952 89 + 1 ---UGCAACUCUGCAGCACUGCAAC-----------------GUUGCAA---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCU ---((((((..((((....))))..-----------------)))))).---..(((((((...(((...((((.......))))((.....))....)))...))))))). ( -29.00) >consensus CAUUGCAACUCUGC___________________A_UGCAAC_UCUGCAA___CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCU ....((((....................................((((.....))))........((.....)).(((((.((((.((.....)).)))).))))).)))). (-16.72 = -16.75 + 0.03)



| Location | 10,249,504 – 10,249,612 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.63 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249504 108 - 27905053 AGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---UUGCAGA-GUUGCAAUGCAGCAGAGUUGCAAUGCCGCAGAGUUGCAAUG .((((((..((.(((((.((..(((((..((.((((.(((.(((.(((.......))).)---)))))))-)).))...)))))...)).)))..)).))..)))))).... ( -37.00) >DroVir_CAF1 124676 85 - 1 AGCAAUUAAGCUGCUGUCAAAUAUUGCCAGCUACUCAUGCUGCUCGAGUUA--GUUACAG---UUGCA---GUUGCC-U--------GUUG--------C-CUGUUGCC-UG .(((((...((.(((((.....((((..((((((((.........))).))--))).)))---).)))---)).)).-.--------))))--------)-........-.. ( -19.40) >DroPse_CAF1 116214 89 - 1 AGCAAUUAAGCCGCUGUCAAAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAGCAGUUGCAACAGUUACA-----------------------ACAGUUGCAAUG .((((((.((((.........))))((((((.......))))...)).....))))))....((((((((.((....-----------------------)).)))))))). ( -26.20) >DroSec_CAF1 107355 92 - 1 AGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---UUGCAGA-GUUGCAAUGC----------------AGCAGAGUUGCAAUG .((((((.((((.........))))((((((.......))))...)).....)))))).(---((((((.-.((((.....----------------.))))..))))))). ( -29.80) >DroSim_CAF1 105815 92 - 1 AGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---UUGCAGA-GUUGCAAUGC----------------CGCAGAGUUGCAAUG .((((((.((((.........))))((((((.......))))...)).....)))))).(---((((((.-.((((.....----------------.))))..))))))). ( -29.80) >DroEre_CAF1 107952 89 - 1 AGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---UUGCAAC-----------------GUUGCAGUGCUGCAGAGUUGCA--- .((((((.((((.........))))((((((.......))))...)).....))))))..---.((((((-----------------.(((((....))))).))))))--- ( -30.00) >consensus AGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG___UUGCAGA_GUUGCA_U___________________GCAGAGUUGCAAUG .((((((.((((.........))))((((((.......))))...(((.......))).......))...................................)))))).... (-16.17 = -16.70 + 0.53)



| Location | 10,249,544 – 10,249,643 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -26.86 |
| Energy contribution | -26.83 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.958003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249544 99 + 27905053 C-UCUG-CAA---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUGUGACAGCGGCUUAAUUGCUGUCUGCAAUUAUGAUUGCUGCC-G-----GUGAAAUG--- .-((.(-((.---..(((((..(((.(((..((((((((((.((((.((.....)).)))).)))))..)))))..)))..)))..)))))))).-)-----).......--- ( -31.60) >DroPse_CAF1 116231 97 + 1 CUGUUG-CAACUGCUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUUUGACAGCGGCUUAAUUGCUGUCUGCAAUUAUGAUUGUUGCC-G-----G------G--- ......-...((((.(((((..(((.(((..((((((((((.((((.(((...))).)))).)))))..)))))..)))..)))..))))).).)-)-----)------.--- ( -31.20) >DroGri_CAF1 112884 99 + 1 C---UGGUAA---CUGCAACUGUAACACGAGCAGCAUGAGUAGCUGGCAAUAUUUGACAGCAGCUUAAUUGCUGUCUGCAAUUAUGAUAGCUGCUUG-----GUGAAAUU--- .---..((.(---(.......)).)).((((((((..((((.((((.((.....)).)))).))))((((((.....))))))......))))))))-----........--- ( -29.80) >DroWil_CAF1 120950 99 + 1 -------UAG---UCGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGACAGCAGUUGACGGCAGCUUAAUUGCUGUCUGCAAUUAUGAUUGUUGCU-GUAUUUGUGAAACU--- -------...---((((((...((((((..(((((.......)))))..)).))))((((((((.(((((((.....))))))).....))))))-))..))))))....--- ( -34.60) >DroAna_CAF1 96826 98 + 1 -----G-UUG---CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGUCUGUGACAGCGGCUUAAUUGCUGUCCGCAAUUAUGAUUGCCUCU-G-----GCGAAGCACCC -----.-(((---(((((((..(((.(((..((((((((((.((((.((.....)).)))).)))))..)))))..)))..)))..)))))....-)-----))))....... ( -34.90) >DroPer_CAF1 118910 84 + 1 --------------UGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUUUGACAGCGGCUUAAUUGCUGUCUGCAAUUAUGAUUGUUGCC-G-----G------G--- --------------.(((((..(((.(((..((((((((((.((((.(((...))).)))).)))))..)))))..)))..)))..)))))....-.-----.------.--- ( -27.20) >consensus _____G_UAA___CUGCAAUUGUAACGCGAUCAGCAUGAGUAGCUGGCAGCCUUUGACAGCGGCUUAAUUGCUGUCUGCAAUUAUGAUUGCUGCC_G_____GUGAAA_G___ ...............((((((((((.(((..((((((((((.((((.((.....)).)))).)))))..)))))..)))..))))))))))...................... (-26.86 = -26.83 + -0.03)



| Location | 10,249,544 – 10,249,643 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -17.76 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10249544 99 - 27905053 ---CAUUUCAC-----C-GGCAGCAAUCAUAAUUGCAGACAGCAAUUAAGCCGCUGUCACAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---UUG-CAGA-G ---........-----.-((((((...((....))..((((((.........))))))....)))))).....(((.(((.(((.(((.......))).)---)))-))))-) ( -28.70) >DroPse_CAF1 116231 97 - 1 ---C------C-----C-GGCAACAAUCAUAAUUGCAGACAGCAAUUAAGCCGCUGUCAAAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAGCAGUUG-CAACAG ---.------.-----.-(....)........((((.((((((.........))))))...((((((((((.......))))...(((.......))).)))))))-)))... ( -28.90) >DroGri_CAF1 112884 99 - 1 ---AAUUUCAC-----CAAGCAGCUAUCAUAAUUGCAGACAGCAAUUAAGCUGCUGUCAAAUAUUGCCAGCUACUCAUGCUGCUCGUGUUACAGUUGCAG---UUACCA---G ---........-----...((((((............(((((((.......))))))).(((((...((((.......))))...)))))..))))))..---......---. ( -25.50) >DroWil_CAF1 120950 99 - 1 ---AGUUUCACAAAUAC-AGCAACAAUCAUAAUUGCAGACAGCAAUUAAGCUGCCGUCAACUGCUGUCAGCUACUCAUGCUGAUCGCGUUACAAUUGCGA---CUA------- ---.............(-((((.......(((((((.....)))))))((((((.((.....)).).))))).....))))).(((((.......)))))---...------- ( -24.90) >DroAna_CAF1 96826 98 - 1 GGGUGCUUCGC-----C-AGAGGCAAUCAUAAUUGCGGACAGCAAUUAAGCCGCUGUCACAGACUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG---CAA-C----- .((((((((..-----.-.))))).....(((((((.....))))))).)))(((((....(((.((((((.......))))...)))))......))))---)..-.----- ( -30.30) >DroPer_CAF1 118910 84 - 1 ---C------C-----C-GGCAACAAUCAUAAUUGCAGACAGCAAUUAAGCCGCUGUCAAAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCA-------------- ---.------.-----.-(((........(((((((.....)))))))((((.........)))))))(((.......)))....(((.......))).-------------- ( -21.20) >consensus ___C_UUUCAC_____C_AGCAACAAUCAUAAUUGCAGACAGCAAUUAAGCCGCUGUCAAAGGCUGCCAGCUACUCAUGCUGAUCGCGUUACAAUUGCAG___UUA_C_____ ...................((((......(((((((.....))))))).((.((.((.....)).))((((.......))))...)).......))))............... (-17.76 = -16.90 + -0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:55 2006