
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 10,245,356 – 10,245,479 |
| Length | 123 |
| Max. P | 0.918604 |

| Location | 10,245,356 – 10,245,450 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -9.50 |
| Energy contribution | -9.37 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850165 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
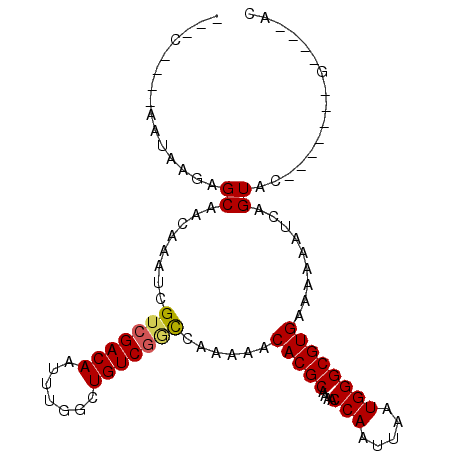
>3R_DroMel_CAF1 10245356 94 + 27905053 AGG-CUCUGC-CAAAAAAUAAAAAGGUUAAC------CUUCUCGAGAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAA .((-(...))-)............((((..(------(((((....))))))((........(((((((.......)))))))((....)).))..)))).. ( -27.30) >DroGri_CAF1 107754 86 + 1 AUG-CUUUAAUCGCAAUGU-GAAAAGUUAA----------C----AAUAUGAGCAACAAAUCGUCGACAAUUUGGCUGUCGACCAAAAACACGCAAAACCAA .((-(.......))).(((-(.........----------.----.....((........))(((((((.......)))))))........))))....... ( -14.20) >DroEre_CAF1 103994 90 + 1 AGG-CUCCGC-CGAA------AAAGGUUAACCCAAACCCAC----GAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAA ..(-(...((-(...------...)))...((((..(((.(----....)))).........(((((((.......))))))))))).....))........ ( -26.00) >DroYak_CAF1 109672 87 + 1 AGG-CUCUGC-CAAAAA---AAAAGGUUAAC------CCAC----GAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAA .((-(...))-).....---....((((..(------((..----....)))((........(((((((.......)))))))((....)).))..)))).. ( -23.90) >DroMoj_CAF1 112186 87 + 1 AGGUUUUUAAUCGCAAUGC-CAAAAGUCAA----------C----AAUAAGAGCAACAAAUCGUCGACAAUUUGGCUGUCGGCCGAAAACACGCAAAACCAA .((((((.....((..(((-(.........----------.----.....).))).....(((((((((.......)))))).)))......)))))))).. ( -15.86) >DroAna_CAF1 93333 85 + 1 -GA-GUGUGU-A-AAAA---AUAAGGUUAAC------ACAG----GAGAGCGGCAACAAAUCGUCGACAAUUUGGCUGUCGCUCGGGAACACGCAAAACCAA -..-((((((-.-....---....(.....)------....----..((((((((.(((((.(....).)))))..))))))))....))))))........ ( -24.00) >consensus AGG_CUCUGC_CGAAAA___AAAAGGUUAAC______CCAC____GAGAGGGGCAACAAAUCGUCGACAAUUUGGCUGUCGACCGGGAACACGCAAAACCAA ....................................................((......(((((((((.......))))))).))......))........ ( -9.50 = -9.37 + -0.14)



| Location | 10,245,356 – 10,245,450 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -10.32 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10245356 94 - 27905053 UUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUCUCGAGAAG------GUUAACCUUUUUAUUUUUUG-GCAGAG-CCU ..((((((((.........((((((((.....))))))))...................((((((------(....)))))))........-))))))-)). ( -29.10) >DroGri_CAF1 107754 86 - 1 UUGGUUUUGCGUGUUUUUGGUCGACAGCCAAAUUGUCGACGAUUUGUUGCUCAUAUU----G----------UUAACUUUUC-ACAUUGCGAUUAAAG-CAU ...((((((..(((...((((((((((.....))))))))((...(((((.......----)----------.))))...))-.))..)))..)))))-).. ( -18.80) >DroEre_CAF1 103994 90 - 1 UUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----GUGGGUUUGGGUUAACCUUU------UUCG-GCGGAG-CCU ..((((((((((...((((((((((((.....)))))))).......((((((....----).)))))))))...))(...------...)-))))))-)). ( -31.40) >DroYak_CAF1 109672 87 - 1 UUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC----GUGG------GUUAACCUUUU---UUUUUG-GCAGAG-CCU ..((((((((.........((((((((.....))))))))......(((((((....----).))------)))).......---......-))))))-)). ( -29.60) >DroMoj_CAF1 112186 87 - 1 UUGGUUUUGCGUGUUUUCGGCCGACAGCCAAAUUGUCGACGAUUUGUUGCUCUUAUU----G----------UUGACUUUUG-GCAUUGCGAUUAAAAACCU ..((((((.((((((.......))).((((((..(((((((((..(.....)..)))----)----------))))).))))-))...)))....)))))). ( -27.10) >DroAna_CAF1 93333 85 - 1 UUGGUUUUGCGUGUUCCCGAGCGACAGCCAAAUUGUCGACGAUUUGUUGCCGCUCUC----CUGU------GUUAACCUUAU---UUUU-U-ACACAC-UC- ..........((((....(((((.((((.((((((....)))))))))).)))))..----..((------(..((......---))..-)-))))))-..- ( -19.90) >consensus UUGGUUUUGCGUGUUCCCAGUCGACAGCCAAAUUGUCGACGAUUUGUAGCCCCUCUC____GUGG______GUUAACCUUUU___UUUUCG_GCAGAG_CCU ..(((...(((.(((....((((((((.....))))))))))).))).)))................................................... (-10.32 = -10.60 + 0.28)



| Location | 10,245,385 – 10,245,479 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 10245385 94 + 27905053 CUUCUCGAGAGAGGGGCUACAAAUCGUCGACAAUUUGGCUGUCGACUGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUAC------CG----AC ..((((....))))((.(((.....(((((((.......)))))))......(((((....(((.....)))))))).........))))------).----.. ( -27.30) >DroVir_CAF1 119619 86 + 1 ---C----AAUAAGAGCAACAAAUUGUCGACAAUUUGGCUGUCAGCCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUUC-------G----UC ---.----.....((((................((((((.....))))))..(((((....(((.....)))))))).........))))-------.----.. ( -20.20) >DroPse_CAF1 111213 94 + 1 CAAC----AGAAACAGCAACAAAUCGUCGACAAUUUGGCUGUCGGCCAGAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUAG------AGUAAAAC ....----.((..............(((((((.......)))))))......(((((....(((.....))))))))......)).....------........ ( -17.30) >DroGri_CAF1 107782 86 + 1 ---C----AAUAUGAGCAACAAAUCGUCGACAAUUUGGCUGUCGACCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUUC-------G----AC ---.----....(((((........(((((((.......)))))))......(((((....(((.....)))))))).........))))-------)----.. ( -22.40) >DroMoj_CAF1 112215 86 + 1 ---C----AAUAAGAGCAACAAAUCGUCGACAAUUUGGCUGUCGGCCGAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUGC-------G----AC ---.----.......(((.....(((((((((.......)))))).)))...(((((....(((.....))))))))..........)))-------.----.. ( -20.80) >DroAna_CAF1 93357 96 + 1 ACAG----GAGAGCGGCAACAAAUCGUCGACAAUUUGGCUGUCGCUCGGGAACACGCAAAACCAAUUAAUGGGCGUGAAAAAGUCAGUACGAGCUCCG----AC ...(----((((((((((.(((((.(....).)))))..)))))))).....(((((....(((.....)))))))).................))).----.. ( -28.70) >consensus ___C____AAUAAGAGCAACAAAUCGUCGACAAUUUGGCUGUCGGCCAAAAACACGCAAAACCAAUUAAUGGGCGUGAAAAAAUCAGUAC_______G____AC ...............((........(((((((.......)))))))......(((((....(((.....)))))))).........))................ (-15.72 = -15.75 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:50 2006